Isoginkgetin, a potential CDK6 inhibitor, suppresses SLC2A1/GLUT1 enhancer activity to induce AMPK-ULK1-mediated cytotoxic autophagy in hepatocellular carcinoma
- PMID: 36048765
- PMCID: PMC10012924
- DOI: 10.1080/15548627.2022.2119353
Isoginkgetin, a potential CDK6 inhibitor, suppresses SLC2A1/GLUT1 enhancer activity to induce AMPK-ULK1-mediated cytotoxic autophagy in hepatocellular carcinoma
Abstract
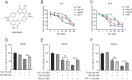
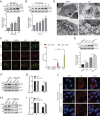
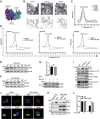
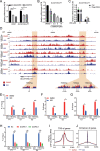
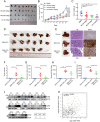
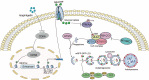
Isoginkgetin (ISO), a natural biflavonoid, exhibited cytotoxic activity against several types of cancer cells. However, its effects on hepatocellular carcinoma (HCC) cells and mechanism remain unclear. Here, we revealed that ISO effectively inhibited HCC cell proliferation and migration in vitro. LC3-II expression and autophagosomes were increased under ISO treatment. In addition, ISO-induced cell death was attenuated by treatment with chloroquine or knockdown of autophagy-related genes (ATG5 or ULK1). ISO significantly suppressed SLC2A1/GLUT1 (solute carrier family 2 member 1) expression and glucose uptake, leading to activation of the AMPK-ULK1 axis in HepG2 cells. Overexpression of SLC2A1/GLUT1 abrogated ISO-induced autophagy. Combining molecular docking with thermal shift analysis, we confirmed that ISO directly bound to the N terminus of CDK6 (cyclin-dependent kinase 6) and promoted its degradation. Overexpression of CDK6 abrogated ISO-induced inhibition of SLC2A1/GLUT1 transcription and induction of autophagy. Furthermore, ISO treatment significantly decreased the H3K27ac, H4K8ac and H3K4me1 levels on the SLC2A1/GLUT1 enhancer in HepG2 cells. Finally, ISO suppressed the hepatocarcinogenesis in the HepG2 xenograft mice and the diethylnitrosamine+carbon tetrachloride (DEN+CCl4)-induced primary HCC mice and we confirmed SLC2A1/GLUT1 and CDK6 as promising oncogenes in HCC by analysis of TCGA data and human HCC tissues. Our results provide a new molecular mechanism by which ISO treatment or CDK6 deletion promotes autophagy; that is, ISO targeting the N terminus of CDK6 for degradation inhibits the expression of SLC2A1/GLUT1 by decreasing the enhancer activity of SLC2A1/GLUT1, resulting in decreased glucose levels and inducing the AMPK-ULK1 pathway.
Keywords: Autophagy; CDK6; SLC2A1/GLUT1; hepatocellular carcinoma; isoginkgetin.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
