Reproductive isolation via polygenic local adaptation in sub-divided populations: Effect of linkage disequilibria and drift
- PMID: 36048903
- PMCID: PMC9473638
- DOI: 10.1371/journal.pgen.1010297
Reproductive isolation via polygenic local adaptation in sub-divided populations: Effect of linkage disequilibria and drift
Abstract
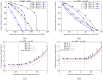
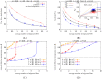
This paper considers how polygenic local adaptation and reproductive isolation between hybridizing populations is influenced by linkage disequilibria (LD) between loci, in scenarios where both gene flow and genetic drift counteract selection. It shows that the combined effects of multi-locus LD and genetic drift on allele frequencies at selected loci and on heterozygosity at neutral loci are predicted accurately by incorporating (deterministic) effective migration rates into the diffusion approximation (for selected loci) and into the structured coalescent (for neutral loci). Theoretical approximations are tested against individual-based simulations and used to investigate conditions for the maintenance of local adaptation on an island subject to one-way migration from a differently adapted mainland, and in an infinite-island population with two habitats under divergent selection. The analysis clarifies the conditions under which LD between sets of locally deleterious alleles allows these to be collectively eliminated despite drift, causing sharper and (under certain conditions) shifted migration thresholds for loss of adaptation. Local adaptation also has counter-intuitive effects on neutral (relative) divergence: FST is highest for a pair of subpopulations belonging to the same (rare) habitat, despite the lack of reproductive isolation between them.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Lind B. M., Menon M., Bolte C. E., Faske T. M., and Eckert A. J. 2018. The genomics of local adaptation in trees: are we out of the woods yet? Tree Genetics and Genomes 14. doi: 10.1007/s11295-017-1224-y - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

