First detection and complete genome analysis of porcine circovirus-like virus P1 and porcine circovirus-2 in yak in China
- PMID: 36049138
- PMCID: PMC9677406
- DOI: 10.1002/vms3.911
First detection and complete genome analysis of porcine circovirus-like virus P1 and porcine circovirus-2 in yak in China
Abstract
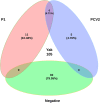
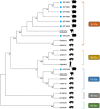
Porcine circovirus-like virus P1, like porcine circovirus type 2 (PCV2), is a potential pathogen of post-weaning multisystemic wasting syndrome in swine. Yaks are a valuable species and an iconic symbol of the Tibet Plateau which is the highest and largest plateau in the world. In this study, a total of 105 yak diarrheal samples, collected from 13 farms in Linzhi in the Tibet Plateau from January 2019 to December 2021, that were screened for P1 and PCV2 by polymerase chain reaction, 10.48% (n = 11) were positive for P1, 4.76% (n = 5) for PCV2, and 5.71% (n = 6) were positive for coinfection of P1 and PCV2. In addition, the whole genomes of eight P1 strains and eight PCV2 strains were sequenced. Alignment of deduced amino acid sequences of P1 ORF1 and PCV2 ORF2 gene revealed that ON012566 had one unique amino acid mutation at residues 137 (T to P). This mutation has important implication for the study of virus virulence, tissue tropism, and immune response. Phylogenetic analysis shows that the yak-origin P1 strains in this study with cattle-origin P1 reference strains were grouped into one cluster. The yak-origin PCV2 (ON012566) and a buffalo-origin PCV2 (KM116514) reference strain clustered in the same branch in the PCV2b regions. Meanwhile, the remaining PCV2 strains and buffalo-origin PCV2 reference strain (ON012565) clustered in the PCV2d regions. To summarize, to our knowledge, this is the first report on the molecular prevalence and genome characteristics of P1 and PCV2 in yaks in the world and will contribute to further study of the molecular epidemiology, source, and evolution of P1 and PCV2 strains.
Keywords: phylogeny; porcine circovirus-2; porcine circovirus-like virus P1; sequencing; yak.
© 2022 The Authors. Veterinary Medicine and Science published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Cheung, A. K. , Lager, K. M. , Kohutyuk, O. I. , Vincent, A. L. , Henry, S. C. , Baker, R. B. , Rowland, R. R. , & Dunham, A. G. (2007). Detection of two porcine circovirus type 2 genotypic groups in United States swine herds. Archives of Virology, 152(5), 1035–1044. 10.1007/s00705-006-0909-6 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

