Photochemical Identification of Auxiliary Severe Acute Respiratory Syndrome Coronavirus 2 Host Entry Factors Using μMap
- PMID: 36049228
- PMCID: PMC9469761
- DOI: 10.1021/jacs.2c06806
Photochemical Identification of Auxiliary Severe Acute Respiratory Syndrome Coronavirus 2 Host Entry Factors Using μMap
Abstract
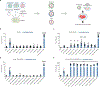
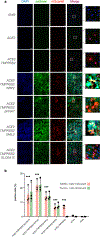
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the infectious agent of the COVID-19 pandemic, remains a global medical problem. Angiotensin-converting enzyme 2 (ACE2) was identified as the primary viral entry receptor, and transmembrane serine protease 2 primes the spike protein for membrane fusion. However, ACE2 expression is generally low and variable across tissues, suggesting that auxiliary receptors facilitate viral entry. Identifying these factors is critical for understanding SARS-Cov-2 pathophysiology and developing new countermeasures. However, profiling host-virus interactomes involves extensive genetic screening or complex computational predictions. Here, we leverage the photocatalytic proximity labeling platform μMap to rapidly profile the spike interactome in human cells and identify eight novel candidate receptors. We systemically validate their functionality in SARS-CoV-2 pseudoviral uptake assays with both Wuhan and Delta spike variants and show that dual expression of ACE2 with either neuropilin-2, ephrin receptor A7, solute carrier family 6 member 15, or myelin and lymphocyte protein 2 significantly enhances viral uptake. Collectively, our data show that SARS-CoV-2 synergistically engages several host factors for cell entry and establishes μMap as a powerful tool for rapidly interrogating host-virus interactomes.
Figures
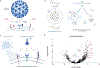
Similar articles
-
Computational Alanine Scanning and Structural Analysis of the SARS-CoV-2 Spike Protein/Angiotensin-Converting Enzyme 2 Complex.ACS Nano. 2020 Sep 22;14(9):11821-11830. doi: 10.1021/acsnano.0c04674. Epub 2020 Aug 26. ACS Nano. 2020. PMID: 32833435 Free PMC article.
-
TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein.J Virol. 2014 Jan;88(2):1293-307. doi: 10.1128/JVI.02202-13. Epub 2013 Nov 13. J Virol. 2014. PMID: 24227843 Free PMC article.
-
Interaction of Human ACE2 to Membrane-Bound SARS-CoV-1 and SARS-CoV-2 S Glycoproteins.Viruses. 2020 Sep 29;12(10):1104. doi: 10.3390/v12101104. Viruses. 2020. PMID: 33003587 Free PMC article.
-
Angiotensin-converting enzyme 2 (ACE2) receptor and SARS-CoV-2: Potential therapeutic targeting.Eur J Pharmacol. 2020 Oct 5;884:173455. doi: 10.1016/j.ejphar.2020.173455. Epub 2020 Jul 31. Eur J Pharmacol. 2020. PMID: 32745604 Free PMC article. Review.
-
Interaction of SARS-CoV-2 and Other Coronavirus With ACE (Angiotensin-Converting Enzyme)-2 as Their Main Receptor: Therapeutic Implications.Hypertension. 2020 Nov;76(5):1339-1349. doi: 10.1161/HYPERTENSIONAHA.120.15256. Epub 2020 Aug 27. Hypertension. 2020. PMID: 32851855 Free PMC article. Review.
Cited by
-
µMap proximity labeling in living cells reveals stress granule disassembly mechanisms.Nat Chem Biol. 2025 Apr;21(4):490-500. doi: 10.1038/s41589-024-01721-2. Epub 2024 Aug 30. Nat Chem Biol. 2025. PMID: 39215100 Free PMC article.
-
Dynamic Covalent Michael Acceptors to Penetrate Cells: Thiol-Mediated Uptake with Tetrel-Centered Exchange Cascades, Assisted by Halogen-Bonding Switches.Angew Chem Int Ed Engl. 2022 Dec 19;61(51):e202213433. doi: 10.1002/anie.202213433. Epub 2022 Nov 17. Angew Chem Int Ed Engl. 2022. PMID: 36272154 Free PMC article.
-
Dynamic Vertical Triplet Energies: Understanding and Predicting Triplet Energy Transfer.Chem. 2024 Nov 14;10(11):3428-3443. doi: 10.1016/j.chempr.2024.07.001. Epub 2024 Jul 26. Chem. 2024. PMID: 39935516
-
Inhibition of Cell Motility by Cell-Penetrating Dynamic Covalent Cascade Exchangers: Integrins Participate in Thiol-Mediated Uptake.JACS Au. 2023 Apr 12;3(4):1010-1016. doi: 10.1021/jacsau.3c00113. eCollection 2023 Apr 24. JACS Au. 2023. PMID: 37124287 Free PMC article.
-
Current advances in photocatalytic proximity labeling.Cell Chem Biol. 2024 Jun 20;31(6):1145-1161. doi: 10.1016/j.chembiol.2024.03.012. Epub 2024 Apr 24. Cell Chem Biol. 2024. PMID: 38663396 Free PMC article. Review.
References
-
- Glowacka I, Bertram S, Müller MA, Allen P, Soilleux E, Pfefferle S, Steffen I, Tsegaye TS, He Y. & Gnirss K. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011, 85 (9), 4122–4134. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

