GSK3β is a critical, druggable component of the network regulating the active NOTCH1 protein and cell viability in CLL
- PMID: 36050315
- PMCID: PMC9436923
- DOI: 10.1038/s41419-022-05178-w
GSK3β is a critical, druggable component of the network regulating the active NOTCH1 protein and cell viability in CLL
Abstract
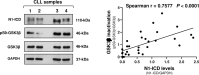
NOTCH1 alterations have been associated with chronic lymphocytic leukemia (CLL), but the molecular mechanisms underlying NOTCH1 activation in CLL cells are not completely understood. Here, we show that GSK3β downregulates the constitutive levels of the active NOTCH1 intracellular domain (N1-ICD) in CLL cells. Indeed, GSK3β silencing by small interfering RNA increases N1-ICD levels, whereas expression of an active GSK3β mutant reduces them. Additionally, the GSK3β inhibitor SB216763 enhances N1-ICD stability at a concentration at which it also increases CLL cell viability. We also show that N1-ICD is physically associated with GSK3β in CLL cells. SB216763 reduces GSK3β/N1-ICD interactions and the levels of ubiquitinated N1-ICD, indicating a reduction in N1-ICD proteasomal degradation when GSK3β is less active. We then modulated the activity of two upstream regulators of GSK3β and examined the impact on N1-ICD levels and CLL cell viability. Specifically, we inhibited AKT that is a negative regulator of GSK3β and is constitutively active in CLL cells. Furthermore, we activated the protein phosphatase 2 A (PP2A) that is a positive regulator of GSK3β, and has an impaired activity in CLL. Results show that either AKT inhibition or PP2A activation reduce N1-ICD expression and CLL cell viability in vitro, through mechanisms mediated by GSK3β activity. Notably, for PP2A activation, we used the highly specific activator DT-061, that also reduces leukemic burden in peripheral blood, spleen and bone marrow in the Eµ-TCL1 adoptive transfer model of CLL, with a concomitant decrease in N1-ICD expression. Overall, we identify in GSK3β a key component of the network regulating N1-ICD stability in CLL, and in AKT and PP2A new druggable targets for disrupting NOTCH1 signaling with therapeutic potential.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

