NRF2 mediates melanoma addiction to GCDH by modulating apoptotic signalling
- PMID: 36050469
- PMCID: PMC9977532
- DOI: 10.1038/s41556-022-00985-x
NRF2 mediates melanoma addiction to GCDH by modulating apoptotic signalling
Abstract
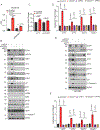
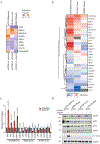
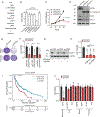
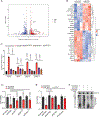
Tumour dependency on specific metabolic signals has been demonstrated and often guided numerous therapeutic approaches. We identify melanoma addiction to the mitochondrial protein glutaryl-CoA dehydrogenase (GCDH), which functions in lysine metabolism and controls protein glutarylation. GCDH knockdown induced cell death programmes in melanoma cells, an activity blocked by inhibition of the upstream lysine catabolism enzyme DHTKD1. The transcription factor NRF2 mediates GCDH-dependent melanoma cell death programmes. Mechanistically, GCDH knockdown induces NRF2 glutarylation, increasing its stability and DNA binding activity, with a concomitant transcriptional upregulation of ATF4, ATF3, DDIT3 and CHAC1, resulting in cell death. In vivo, inducible inactivation of GCDH effectively inhibited melanoma tumour growth. Correspondingly, reduced GCDH expression correlated with improved survival of patients with melanoma. These findings identify melanoma cell addiction to GCDH, limiting apoptotic signalling by controlling NRF2 glutarylation. Inhibiting the GCDH pathway could thus represent a therapeutic approach to treat melanoma.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Interests Statement
ZAR and ER (fully divested) are founders and scientific advisors for Pangea Therapeutics. All other authors declare no conflict of interest.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

