Interplay of transcription factors orchestrating the biosynthesis of plant alkaloids
- PMID: 36051988
- PMCID: PMC9424429
- DOI: 10.1007/s13205-022-03316-x
Interplay of transcription factors orchestrating the biosynthesis of plant alkaloids
Abstract
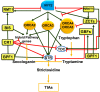
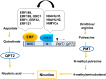
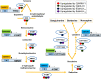
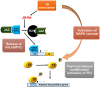
Plants produce a range of secondary metabolites primarily as defence molecules. A plant has to invest considerable energy to synthesise alkaloids, and sometimes they are even toxic to themselves. Hence, the biosynthesis of alkaloids is a spatiotemporally regulated process under quantitative feedback regulation which is accomplished by the signal reception, transcriptional/translational regulation, transport, storage and accumulation. The transcription factors (TFs) initiate the biosynthesis of alkaloids after appropriate cues. The present study recapitulates last decade understanding of the role of TFs in alkaloid biosynthesis. The present review discusses TF families, viz. AP2/ERF, bHLH, WRKY, MYB involved in the biosynthesis of various types of alkaloids. It also highlights the role of the jasmonic acid cascade and post-translational modifications of TF proteins. A thorough understanding of TFs will help us to decide a strategy to facilitate successful pathway manipulation and in vitro production.
Keywords: AP2/ERF; JA; Regulation; Transcription factors.
© King Abdulaziz City for Science and Technology 2022, Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestOn behalf of all authors, the corresponding author states that there is no conflict of interest. The authors have no relevant financial or non-financial interests to disclose.
Figures
Similar articles
-
Phylogenomics of transcriptionally active AP2/ERF and bHLH transcription factors and study of their promoter regions in Nothapodytes nimmoniana (J.Graham) Mabb.Genome. 2023 Sep 1;66(9):235-250. doi: 10.1139/gen-2023-0009. Epub 2023 May 10. Genome. 2023. PMID: 37163758
-
Transcription factors in alkaloid biosynthesis.Int Rev Cell Mol Biol. 2013;305:339-82. doi: 10.1016/B978-0-12-407695-2.00008-1. Int Rev Cell Mol Biol. 2013. PMID: 23890386 Review.
-
A differentially regulated AP2/ERF transcription factor gene cluster acts downstream of a MAP kinase cascade to modulate terpenoid indole alkaloid biosynthesis in Catharanthus roseus.New Phytol. 2017 Feb;213(3):1107-1123. doi: 10.1111/nph.14252. Epub 2016 Nov 1. New Phytol. 2017. PMID: 27801944
-
AP2/ERF, an important cold stress-related transcription factor family in plants: A review.Physiol Mol Biol Plants. 2021 Sep;27(9):1953-1968. doi: 10.1007/s12298-021-01061-8. Epub 2021 Sep 13. Physiol Mol Biol Plants. 2021. PMID: 34616115 Free PMC article. Review.
-
Transcriptional cascades in the regulation of 2-AP biosynthesis under Zn supply in fragrant rice.Physiol Plant. 2022 May;174(3):e13721. doi: 10.1111/ppl.13721. Physiol Plant. 2022. PMID: 35598224
Cited by
-
Genome-wide analysis of the ERF Family in Stephania japonica provides insights into the regulatory role in Cepharanthine biosynthesis.Front Plant Sci. 2024 Sep 4;15:1433015. doi: 10.3389/fpls.2024.1433015. eCollection 2024. Front Plant Sci. 2024. PMID: 39297007 Free PMC article.
-
Regulation of Steroidal Alkaloid Biosynthesis in Bulbs of Fritillaria thunbergii Miq. By Shading and Potassium Application: Integrating Transcriptomics and Metabolomics Analyses.Biology (Basel). 2025 May 29;14(6):633. doi: 10.3390/biology14060633. Biology (Basel). 2025. PMID: 40563884 Free PMC article.
-
Time-Series Metabolome and Transcriptome Analyses Reveal the Genetic Basis of Vanillin Biosynthesis in Vanilla.Plants (Basel). 2025 Jun 23;14(13):1922. doi: 10.3390/plants14131922. Plants (Basel). 2025. PMID: 40647930 Free PMC article.
-
Transcriptomic and metabolomic analyses provide new insights into the appropriate harvest period in regenerated bulbs of Fritillaria hupehensis.Front Plant Sci. 2023 Feb 15;14:1132936. doi: 10.3389/fpls.2023.1132936. eCollection 2023. Front Plant Sci. 2023. PMID: 36875619 Free PMC article.
-
Network-based identification of hub transcription factors associated with benzylisoquinoline alkaloid biosynthesis in Papaver somniferum.Biochem Biophys Rep. 2025 Jul 23;43:102147. doi: 10.1016/j.bbrep.2025.102147. eCollection 2025 Sep. Biochem Biophys Rep. 2025. PMID: 40740472 Free PMC article.
References
-
- Afrin S, Huang JJ, Luo ZY. JA-mediated transcriptional regulation of secondary metabolism in medicinal plants. Sci Bull. 2015;60:1062–1072. doi: 10.1007/s11434-015-0813-0. - DOI
-
- Ali AH, Abdelrahman M, El-Sayed MA. Bioactive molecules in plant defense: signaling in growth and stress. Cham: Springer; 2019. Alkaloid role in plant defense response to growth and stress; pp. 145–158.
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

