Erythropoietin receptor regulates tumor mitochondrial biogenesis through iNOS and pAKT
- PMID: 36052260
- PMCID: PMC9425774
- DOI: 10.3389/fonc.2022.976961
Erythropoietin receptor regulates tumor mitochondrial biogenesis through iNOS and pAKT
Abstract
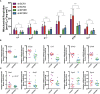
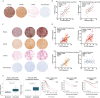
Erythropoietin receptor (EPOR) is widely expressed in healthy and malignant tissues. In certain malignancies, EPOR stimulates tumor growth. In healthy tissues, EPOR controls processes other than erythropoiesis, including mitochondrial metabolism. We hypothesized that EPOR also controls the mitochondrial metabolism in cancer cells. To test this hypothesis, we generated EPOR-knockdown cancer cells to grow tumor xenografts in mice and analyzed tumor cellular respiration via high-resolution respirometry. Furthermore, we analyzed cellular respiratory control, mitochondrial content, and regulators of mitochondrial biogenesis in vivo and in vitro in different cancer cell lines. Our results show that EPOR controls tumor growth and mitochondrial biogenesis in tumors by controlling the levels of both, pAKT and inducible NO synthase (iNOS). Furthermore, we observed that the expression of EPOR is associated with the expression of the mitochondrial marker VDAC1 in tissue arrays of lung cancer patients, suggesting that EPOR indeed helps to regulate mitochondrial biogenesis in tumors of cancer patients. Thus, our data imply that EPOR not only stimulates tumor growth but also regulates tumor metabolism and is a target for direct intervention against progression.
Keywords: OXPHOS; VDAC1; erythropoietin receptor; mitochondrial biogenesis; nitric oxide (NO); respirometry; tumor metabolism.
Copyright © 2022 Aboouf, Guscetti, von Büren, Armbruster, Ademi, Ruetten, Meléndez-Rodríguez, Rülicke, Seymer, Jacobs, Schneider Gasser, Aragones, Neumann, Gassmann and Thiersch.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Tankiewicz-Kwedlo A, Hermanowicz J, Surazynski A, Rozkiewicz D, Pryczynicz A, Domaniewski T, et al. Erythropoietin accelerates tumor growth through increase of erythropoietin receptor (Epor) as well as by the stimulation of angiogenesis in dld-1 and ht-29 xenografts. Mol Cell Biochem (2016) 421(1-2):1–18. doi: 10.1007/s11010-016-2779-x - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

