A Novel Defined RAS-Related Gene Signature for Predicting the Prognosis and Characterization of Biological Function in Osteosarcoma
- PMID: 36052285
- PMCID: PMC9427258
- DOI: 10.1155/2022/5939158
A Novel Defined RAS-Related Gene Signature for Predicting the Prognosis and Characterization of Biological Function in Osteosarcoma
Abstract
Background: Osteosarcoma (OS) is the most common primary bone malignancy in children and adolescents with a high incidence and poor prognosis. Activation of the RAS pathway promotes progression and metastasis of osteosarcoma. RAS has been studied in many different tumors; however, the prognostic value of RAS-associated genes in OS remains unclear. On this basis, we investigated the RAS-related gene signature and explored the intrinsic biological features of OS.
Methods: We obtained RNA transcriptome sequencing data and clinical information of osteosarcoma patients from the TARGET database. RAS pathway-related genes were obtained from the KEGG pathway database. Molecular subgroups and risk models were developed using consensus clustering and least absolute shrinkage and selection operator (LASSO) regression, respectively. ESTIMATE algorithm and ssGSEA analysis were used to assess the tumor microenvironment and immune penetrance between the two groups. A comprehensive review of gene ontology (GO) and KEGG analyses revealed inherent biological functional differences between the two groups.
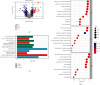
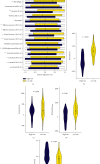
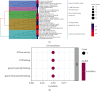
Results: The consistent clustering showed stratification of osteosarcoma patients into two subtypes based on RAS-associated genes and provided a robust prediction of prognosis. A risk model further confirmed that RAS-related genes are the best prognostic indicators for OS patients. GO analysis showed that GDP/GTP binding, focal adhesion, cytoskeletal motor activity, and cell-matrix junctions were associated with the RAS-related model group. Furthermore, RAS signaling in osteosarcoma based on KEGG analysis was significantly associated with cancer progression, with immune function and tumor microenvironment particularly affected.
Conclusion: We constructed a prognostic model founded on RAS-related gene and demonstrated its predictive ability. Then, furtherly exploration of the molecular mechanisms and immune characteristics proved the role of RAS-related gene in the dysregulation in OS.
Copyright © 2022 Qin Chen et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



References
-
- Ottaviani G., Jaffe N. The epidemiology of osteosarcoma. Cancer Treatment and Research . 2009;152:3–13. - PubMed
LinkOut - more resources
Full Text Sources

