Umbilical cord plasma concentrate has beneficial effects on DNA methylation GrimAge and human clinical biomarkers
- PMID: 36052758
- PMCID: PMC9577957
- DOI: 10.1111/acel.13696
Umbilical cord plasma concentrate has beneficial effects on DNA methylation GrimAge and human clinical biomarkers
Abstract
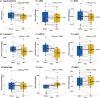
Plasma transfusions are standard treatments to replace missing proteins in people with rare genetic diseases. Prior studies have demonstrated that heterochronic parabiosis has beneficial effects on several tissues of old animals receiving young blood. Human clinical trials are currently underway to investigate whether the infusion of plasma or plasma-derived factors from young donors can be used to mitigate human age-related conditions. Here, we use data from a safety study (n = 18, mean age 74) to investigate whether human umbilical cord plasma concentrate (hereinafter Plasma Concentrate) injected weekly (1 ml intramuscular) into elderly human subjects over a 10-week period affects different biomarkers, including epigenetic age measures, standard clinical biomarkers of organ dysfunction, mitochondrial DNA copy number (mtDNA-CN), and leukocyte telomere length. This study shows that treatment with plasma concentrate is safe. More than 20 clinical biomarkers were significantly and beneficially altered following the treatments. For example, creatinine was significantly decreased (p = 0.0039), while estimated glomerular filtration rate (eGFR) was increased (p = 0.0044), indicating the treatment may improve biomarkers of kidney function. Three of four immunoglobulin biomarkers decreased, while telomere length and mtDNA-CN were not significantly affected by the treatment. The treatment reduced DNA methylation-based GrimAge by an average of 0.82 years (p = 0.0093), suggests a reduction in morbidity and mortality risk. By contrast, no significant results could be observed for epigenetic clocks that estimate chronological age. Our results support the view that plasma concentrate contains youth-promoting factors.
Keywords: clinical trial; epigenetic clocks; exosome treatment; umbilical cord plasma; young plasma.
© 2022 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
James Clement is a founder of Betterhumans Inc., a nonprofit medical research organization with offices in Gainesville, FL and Tyler, Texas focused on slowing and reversing the detrimental effects of aging. Robert T. Brooke and Steve Horvath are founders of the nonprofit Epigenetic Clock Development Foundation. The Clock foundation licenses inventions and patents surrounding epigenetic clocks from UC Regents. These patents list Steve Horvath as inventor.
Figures




References
-
- Castellano, J. M. , Mosher, K. I. , Abbey, R. J. , Mcbride, A. A. , James, M. L. , Berdnik, D. , Shen, J. C. , Zou, B. , Xie, X. S. , Tingle, M. , Hinkson, I. V. , Angst, M. S. , & Wyss‐Coray, T. (2017). Human umbilical cord plasma proteins revitalize hippocampal function in aged mice. Nature, 544, 488–492. - PMC - PubMed
-
- Conboy, I. M. , Conboy, M. J. , Wagers, A. J. , Girma, E. R. , Weissman, I. L. , & Rando, T. A. (2005). Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature, 433, 760–764. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

