Origin of cytoplasmic GDP-fucose determines its contribution to glycosylation reactions
- PMID: 36053214
- PMCID: PMC9441714
- DOI: 10.1083/jcb.202205038
Origin of cytoplasmic GDP-fucose determines its contribution to glycosylation reactions
Abstract
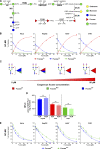
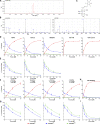
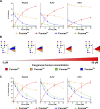
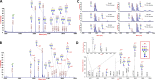
Biosynthesis of macromolecules requires precursors such as sugars or amino acids, originating from exogenous/dietary sources, reutilization/salvage of degraded molecules, or de novo synthesis. Since these sources are assumed to contribute to one homogenous pool, their individual contributions are often overlooked. Protein glycosylation uses monosaccharides from all the above sources to produce nucleotide sugars required to assemble hundreds of distinct glycans. Here, we demonstrate that cells identify the origin/heritage of the monosaccharide, fucose, for glycosylation. We measured the contribution of GDP-fucose from each of these sources for glycan synthesis and found that different fucosyltransferases, individual glycoproteins, and linkage-specific fucose residues identify and select different GDP-fucose pools dependent on their heritage. This supports the hypothesis that GDP-fucose exists in multiple, distinct pools, not as a single homogenous pool. The selection is tightly regulated since the overall pool size remains constant. We present novel perspectives on monosaccharide metabolism, which may have a general applicability.
© 2022 Sosicka et al.
Figures







References
-
- Allen, J.G., Mujacic M., Frohn M.J., Pickrell A.J., Kodama P., Bagal D., San Miguel T., Sickmier E.A., Osgood S., Swietlow A., et al. . 2016. Facile modulation of antibody fucosylation with small molecule fucostatin inhibitors and cocrystal structure with GDP-mannose 4, 6-dehydratase. ACS Chem. Biol. 11:2734–2743. 10.1021/acschembio.6b00460 - DOI - PubMed
-
- Aula, N., Salomäki P., Timonen R., Verheijen F., Mancini G., Månsson J.E., Aula P., and Peltonen L.. 2000. The spectrum of SLC17A5-gene mutations resulting in free sialic acid-storage diseases indicates some genotype-phenotype correlation. Am. J. Hum. Gen. 67:832–840. 10.1086/303077 - DOI - PMC - PubMed
-
- Benz, B.A., Nandadasa S., Takeuchi M., Grady R.C., Takeuchi H., LoPilato R.K., Kakuda S., Somerville R.P.T., Apte S.S., Haltiwanger R.S., and Holdener B.C.. 2016. Genetic and biochemical evidence that gastrulation defects in Pofut2 mutants result from defects in ADAMTS9 secretion. Dev. Biol. 416:111–122. 10.1016/j.ydbio.2016.05.038 - DOI - PMC - PubMed

