Chronic activation of pDCs in autoimmunity is linked to dysregulated ER stress and metabolic responses
- PMID: 36053251
- PMCID: PMC9441715
- DOI: 10.1084/jem.20221085
Chronic activation of pDCs in autoimmunity is linked to dysregulated ER stress and metabolic responses
Abstract
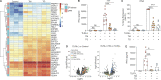
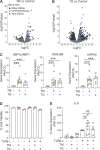
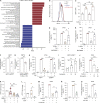
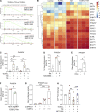
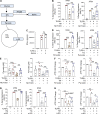
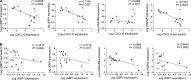
Plasmacytoid dendritic cells (pDCs) chronically produce type I interferon (IFN-I) in autoimmune diseases, including systemic sclerosis (SSc) and systemic lupus erythematosus (SLE). We report that the IRE1α-XBP1 branch of the unfolded protein response (UPR) inhibits IFN-α production by TLR7- or TLR9-activated pDCs. In SSc patients, UPR gene expression was reduced in pDCs, which inversely correlated with IFN-I-stimulated gene expression. CXCL4, a chemokine highly secreted in SSc patients, downregulated IRE1α-XBP1-controlled genes and promoted IFN-α production by pDCs. Mechanistically, IRE1α-XBP1 activation rewired glycolysis to serine biosynthesis by inducing phosphoglycerate dehydrogenase (PHGDH) expression. This process reduced pyruvate access to the tricarboxylic acid (TCA) cycle and blunted mitochondrial ATP generation, which are essential for pDC IFN-I responses. Notably, PHGDH expression was reduced in pDCs from patients with SSc and SLE, and pharmacological blockade of TCA cycle reactions inhibited IFN-I responses in pDCs from these patients. Hence, modulating the IRE1α-XBP1-PHGDH axis may represent a hitherto unexplored strategy for alleviating chronic pDC activation in autoimmune disorders.
© 2022 Chaudhary et al.
Conflict of interest statement
Disclosures: R.L. Wiseman reported “other” from Protego Biopharma outside the submitted work; in addition, R.L. Wiseman had a patent to US20210008064 pending; and is a shareholder and scientific advisory board member of Protego Biopharma, who are developing UPR activating compounds including IXA4. M.K. Crow reported personal fees from AMPEL Biosolutions, BMS, AstraZeneca, GSK, Lilly, and Shannon Pharmaceuticals and grants from Gilead outside the submitted work; in addition, M.K. Crow had a patent number 9,809,854 issued. J.R. Cubillos-Ruiz reported “other” from Quentis Therapeutics, Inc., NextRNA Therapeutics, Inc., Autoimmunity Biologic Solutions, and Immagene, B.V. outside the submitted work; in addition, J.R. Cubillos-Ruiz had a patent to Small molecule IRE1-α inhibitors US10988461B2 issued. F.J. Barrat reported “other” from IpiNovyx Bio outside the submitted work; in addition, F.J. Barrat had a patent to CXCL4 in autoimmunity pending. No other disclosures were reported.
Figures





References
-
- Adamson, B., Norman T.M., Jost M., Cho M.Y., Nunez J.K., Chen Y., Villalta J.E., Gilbert L.A., Horlbeck M.A., Hein M.Y., et al. . 2016. A multiplexed single-cell CRISPR screening Platform enables systematic dissection of the unfolded protein response. Cell. 167:1867–1882.e21. 10.1016/j.cell.2016.11.048 - DOI - PMC - PubMed
-
- Affandi, A.J., Carvalheiro T., Ottria A., de Haan J.J., Brans M.A.D., Brandt M.M., Tieland R.G., Lopes A.P., Fernandez B.M., Bekker C.P.J., et al. . 2022. CXCL4 drives fibrosis by promoting several key cellular and molecular processes. Cell Rep. 38:110189. 10.1016/j.celrep.2021.110189 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

