Megakaryopoiesis impairment through acute innate immune signaling activation by azacitidine
- PMID: 36053753
- PMCID: PMC9441716
- DOI: 10.1084/jem.20212228
Megakaryopoiesis impairment through acute innate immune signaling activation by azacitidine
Abstract
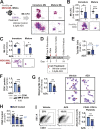
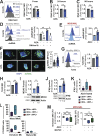
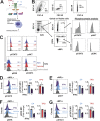
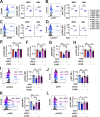
Thrombocytopenia, prevalent in the majority of patients with myeloid malignancies, such as myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML), is an independent adverse prognostic factor. Azacitidine (AZA), a mainstay therapeutic agent for stem cell transplant-ineligible patients with MDS/AML, often transiently induces or further aggravates disease-associated thrombocytopenia by an unknown mechanism. Here, we uncover the critical role of an acute type-I interferon (IFN-I) signaling activation in suppressing megakaryopoiesis in AZA-mediated thrombocytopenia. We demonstrate that megakaryocytic lineage-primed progenitors present IFN-I receptors and, upon AZA exposure, engage STAT1/SOCS1-dependent downstream signaling prematurely attenuating thrombopoietin receptor (TPO-R) signaling and constraining megakaryocytic progenitor cell growth and differentiation following TPO-R stimulation. Our findings directly implicate RNA demethylation and IFN-I signal activation as a root cause for AZA-mediated thrombocytopenia and suggest mitigation of TPO-R inhibitory innate immune signaling as a suitable therapeutic strategy to support platelet production, particularly during the early phases of AZA therapy.
© 2022 Okoye-Okafor et al.
Conflict of interest statement
Disclosures: C. Pallaud reported “other” from Novartis Pharmaceuticals during the conduct of the study. P.M. Ramos is an employee of Novartis Pharmaceuticals. A. Shastri reported grants from Kymera Therapeutics, personal fees from Janssen Pharmaceuticals, and “other” from NACE outside the submitted work. A. Verma reported “other” from Stelexis, Throws Exception, and Bakx Therapeutics; and grants from Curis, Prelude, and BMS outside the submitted work. C. Heckman reported grants from Novartis Pharmaceuticals during the conduct of the study; and grants from BMS/Celgene, Kronos Bio, Oncopeptides, Orion Pharma, Innovative Medicines Initiative Joint Undertaking project HARMONY, and WntResearch outside the submitted work. B. Will reported grants from NIH-NCI, NIH-NIDDK, GlaxoSmithKline, and Novartis; and personal fees from Novartis during the conduct of the study. No other disclosures were reported.
Figures






References
-
- Abramovich, C., Shulman L.M., Ratovitski E., Harroch S., Tovey M., Eid P., and Revel M.. 1994. Differential tyrosine phosphorylation of the IFNAR chain of the type I interferon receptor and of an associated surface protein in response to IFN-alpha and IFN-beta. EMBO J. 13:5871–5877. 10.1002/j.1460-2075.1994.tb06932.x - DOI - PMC - PubMed
-
- Aparicio, A., and Weber J.S.. 2002. Review of the clinical experience with 5-azacytidine and 5-aza-2'-deoxycytidine in solid tumors. Curr. Opin. Investig. Drugs. 3:627–633 - PubMed
-
- Bachegowda, L., Morrone K., Winski S.L., Mantzaris I., Bartenstein M., Ramachandra N., Giricz O., Sukrithan V., Nwankwo G., Shahnaz S., et al. . 2016. Pexmetinib: A novel dual inhibitor of Tie2 and p38 MAPK with efficacy in preclinical models of myelodysplastic syndromes and acute myeloid leukemia. Cancer Res. 76:4841–4849. 10.1158/0008-5472.CAN-15-3062 - DOI - PMC - PubMed
-
- Benjamin, R., Khwaja A., Singh N., McIntosh J., Meager A., Wadhwa M., Streck C., Ng C., Davidoff A.M., and Nathwani A.C.. 2007. Continuous delivery of human type I interferons (alpha/beta) has significant activity against acute myeloid leukemia cells in vitro and in a xenograft model. Blood. 109:1244–1247. 10.1182/blood-2006-02-002915 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

