Characterization of Extracellular Vesicles in Osteoporotic Patients Compared to Osteopenic and Healthy Controls
- PMID: 36053959
- PMCID: PMC10086946
- DOI: 10.1002/jbmr.4688
Characterization of Extracellular Vesicles in Osteoporotic Patients Compared to Osteopenic and Healthy Controls
Abstract
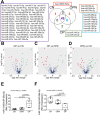
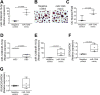
Extracellular vesicles (EVs) are mediators of a range of pathological conditions. However, their role in bone loss disease has not been well understood. In this study we characterized plasma EVs of 54 osteoporotic (OP) postmenopausal women compared to 48 osteopenic (OPN) and 44 healthy controls (CN), and we investigated their effects on osteoclasts and osteoblasts. We found no differences between the three groups in terms of anthropometric measurements and biochemical evaluation of serum calcium, phosphate, creatinine, PTH, 25-hydroxy vitamin D and bone biomarkers, except for an increase of CTX level in OP group. FACS analysis revealed that OP patients presented a significantly increased number of EVs and RANKL+ EVs compared with both CN and OPN subjects. Total EVs are negatively associated with the lumbar spine T-score and femoral neck T-score. Only in the OPN patients we observed a positive association between the total number of EVs and RANKL+ EVs with the serum RANKL. In vitro studies revealed that OP EVs supported osteoclastogenesis of healthy donor peripheral blood mononuclear cells at the same level observed following RANKL and M-CSF treatment, reduced the ability of mesenchymal stem cells to differentiate into osteoblasts, while inducing an increase of OSTERIX and RANKL expression in mature osteoblasts. The analysis of miRNome revealed that miR-1246 and miR-1224-5p were the most upregulated and downregulated in OP EVs; the modulated EV-miRNAs in OP and OPN compared to CN are related to osteoclast differentiation, interleukin-13 production and regulation of canonical WNT pathway. A proteomic comparison between OPN and CN EVs evidenced a decrease in fibrinogen, vitronectin, and clusterin and an increase in coagulation factors and apolipoprotein, which was also upregulated in OP EVs. Interestingly, an increase in RANKL+ EVs and exosomal miR-1246 was also observed in samples from patients affected by Gorham-Stout disease, suggesting that EVs could be good candidate as bone loss disease biomarkers. © 2022 The Authors. Journal of Bone and Mineral Research published by Wiley Periodicals LLC on behalf of American Society for Bone and Mineral Research (ASBMR).
Keywords: MICROVESICLES; MIRNA; OSTEOCLAST; OSTEOPOROSIS; PROTEOMIC ANALYSIS.
© 2022 The Authors. Journal of Bone and Mineral Research published by Wiley Periodicals LLC on behalf of American Society for Bone and Mineral Research (ASBMR).
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19(2):43‐51. - PubMed
-
- Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73(10):1907‐1920. - PubMed
-
- van der Pol E, Boing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64(3):676‐705. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

