The immunosuppression pathway of tumor-associated macrophages is controlled by heme oxygenase-1 in glioblastoma patients
- PMID: 36054818
- PMCID: PMC9825884
- DOI: 10.1002/ijc.34270
The immunosuppression pathway of tumor-associated macrophages is controlled by heme oxygenase-1 in glioblastoma patients
Abstract
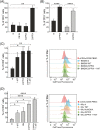
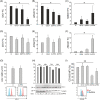
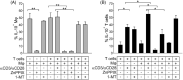
The immunosuppressive tumor microenvironment (TME) in glioblastoma (GBM) is mainly driven by tumor-associated macrophages (TAMs). We explored whether their sustained iron metabolism and immunosuppressive activity were correlated, and whether blocking the central enzyme of the heme catabolism pathway, heme oxygenase-1 (HO-1), could reverse their tolerogenic activity. To this end, we investigated iron metabolism in bone marrow-derived macrophages (BMDMs) isolated from GBM specimens and in in vitro-derived macrophages (Mφ) from healthy donor (HD) blood monocytes. We found that HO-1 inhibition abrogated the immunosuppressive activity of both BMDMs and Mφ, and that immunosuppression requires both cell-to-cell contact and soluble factors, as HO-1 inhibition abolished IL-10 release, and significantly reduced STAT3 activation as well as PD-L1 expression. Interestingly, not only did HO-1 inhibition downregulate IDO1 and ARG-2 gene expression, but also reduced IDO1 enzymatic activity. Moreover, T cell activation status affected PD-L1 expression and IDO1 activity, which were upregulated in the presence of activated, but not resting, T cells. Our results highlight the crucial role of HO-1 in the immunosuppressive activity of macrophages in the GBM TME and demonstrate the feasibility of reprogramming them as an alternative therapeutic strategy for restoring immune surveillance.
Keywords: glioblastoma; heme oxygenase-1; iron metabolism; macrophages; tumor microenvironment.
© 2022 The Authors. International Journal of Cancer published by John Wiley & Sons Ltd on behalf of UICC.
Conflict of interest statement
The authors report that there are no competing interests to declare.
Figures



References
-
- Bejarano L, Jordao MJC, Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Discov. 2021;11(4):933‐959. doi: 10.1158/2159-8290.CD-20-1808 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

