Modeling human extraembryonic mesoderm cells using naive pluripotent stem cells
- PMID: 36055191
- PMCID: PMC9438972
- DOI: 10.1016/j.stem.2022.08.001
Modeling human extraembryonic mesoderm cells using naive pluripotent stem cells
Abstract
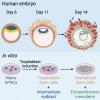
A hallmark of primate postimplantation embryogenesis is the specification of extraembryonic mesoderm (EXM) before gastrulation, in contrast to rodents where this tissue is formed only after gastrulation. Here, we discover that naive human pluripotent stem cells (hPSCs) are competent to differentiate into EXM cells (EXMCs). EXMCs are specified by inhibition of Nodal signaling and GSK3B, are maintained by mTOR and BMP4 signaling activity, and their transcriptome and epigenome closely resemble that of human and monkey embryo EXM. EXMCs are mesenchymal, can arise from an epiblast intermediate, and are capable of self-renewal. Thus, EXMCs arising via primate-specific specification between implantation and gastrulation can be modeled in vitro. We also find that most of the rare off-target cells within human blastoids formed by triple inhibition (Kagawa et al., 2021) correspond to EXMCs. Our study impacts our ability to model and study the molecular mechanisms of early human embryogenesis and related defects.
Keywords: extraembryonic mesoderm; human blastoids; human embryos; human naive pluripotent stem cells.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The Institute for Molecular Biotechnology, Austrian Academy of Sciences has filed patent application EP21151455.9 describing the protocols for human blastoid formation. H.K. and N.R. are the inventors of this patent. All other authors declare no competing interests. J.R. is a member of the Cell Stem Cell advisory board.
Figures






Comment in
-
Exploring the human extraembryonic mesoderm using naive pluripotent stem cells.Cell Stem Cell. 2022 Sep 1;29(9):1290-1291. doi: 10.1016/j.stem.2022.08.005. Cell Stem Cell. 2022. PMID: 36055188
References
-
- Arora S., Morgan M., Carlson M., Pagès H. 2022. GenomeInfoDb: Utilities for manipulating chromosome names, including modifying them to follow a particular naming style.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

