High-coverage whole-genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios
- PMID: 36055201
- PMCID: PMC9439720
- DOI: 10.1016/j.cell.2022.08.004
High-coverage whole-genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios
Abstract
The 1000 Genomes Project (1kGP) is the largest fully open resource of whole-genome sequencing (WGS) data consented for public distribution without access or use restrictions. The final, phase 3 release of the 1kGP included 2,504 unrelated samples from 26 populations and was based primarily on low-coverage WGS. Here, we present a high-coverage 3,202-sample WGS 1kGP resource, which now includes 602 complete trios, sequenced to a depth of 30X using Illumina. We performed single-nucleotide variant (SNV) and short insertion and deletion (INDEL) discovery and generated a comprehensive set of structural variants (SVs) by integrating multiple analytic methods through a machine learning model. We show gains in sensitivity and precision of variant calls compared to phase 3, especially among rare SNVs as well as INDELs and SVs spanning frequency spectrum. We also generated an improved reference imputation panel, making variants discovered here accessible for association studies.
Keywords: 1000 Genomes Project; INDEL; SNV; population genetics; reference imputation panel; structural variation; trio sequencing; whole-genome sequencing.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests E.E.E. is a scientific advisory board (SAB) member of Variant Bio, Inc. P.F. is an SAB member of Fabric Genomics, Inc., and Eagle Genomics, Ltd.
Figures




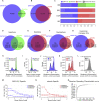
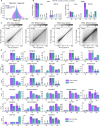


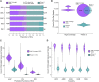
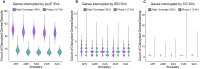

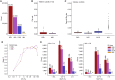

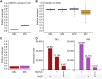
Comment in
-
1000 Genomes Project phase 4: The gift that keeps on giving.Cell. 2022 Sep 1;185(18):3286-3289. doi: 10.1016/j.cell.2022.08.001. Cell. 2022. PMID: 36055197
References
-
- Almeida R., Ricaño-Ponce I., Kumar V., Deelen P., Szperl A., Trynka G., Gutierrez-Achury J., Kanterakis A., Westra H.-J., Franke L., et al. Fine mapping of the celiac disease-associated LPP locus reveals a potential functional variant. Hum. Mol. Genet. 2014;23:2481–2489. doi: 10.1093/hmg/ddt619. - DOI - PMC - PubMed
-
- Andrews S. FastQC. 2019. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Broad Institute Picard Toolkit, Github Repository. 2019. http://broadinstitute.github.io/picard/
Publication types
MeSH terms
Grants and funding
- R01 HG002898/HG/NHGRI NIH HHS/United States
- R03 HD099547/HD/NICHD NIH HHS/United States
- R35 GM138212/GM/NIGMS NIH HHS/United States
- UM1 HG008895/HG/NHGRI NIH HHS/United States
- UM1 HG008901/HG/NHGRI NIH HHS/United States
- R01 HD081256/HD/NICHD NIH HHS/United States
- R56 MH115957/MH/NIMH NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- U24 HG007497/HG/NHGRI NIH HHS/United States
- R21 CA259309/CA/NCI NIH HHS/United States
- R01 MH115957/MH/NIMH NIH HHS/United States
- R01 CA261934/CA/NCI NIH HHS/United States
- UM1 HG008853/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

