Regulation of membrane protein structure and function by their lipid nano-environment
- PMID: 36056103
- PMCID: PMC9892264
- DOI: 10.1038/s41580-022-00524-4
Regulation of membrane protein structure and function by their lipid nano-environment
Erratum in
-
Author Correction: Regulation of membrane protein structure and function by their lipid nano-environment.Nat Rev Mol Cell Biol. 2023 Jan;24(1):79. doi: 10.1038/s41580-022-00560-0. Nat Rev Mol Cell Biol. 2023. PMID: 36329215 No abstract available.
Abstract
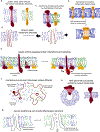
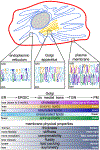
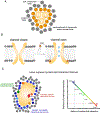
Transmembrane proteins comprise ~30% of the mammalian proteome, mediating metabolism, signalling, transport and many other functions required for cellular life. The microenvironment of integral membrane proteins (IMPs) is intrinsically different from that of cytoplasmic proteins, with IMPs solvated by a compositionally and biophysically complex lipid matrix. These solvating lipids affect protein structure and function in a variety of ways, from stereospecific, high-affinity protein-lipid interactions to modulation by bulk membrane properties. Specific examples of functional modulation of IMPs by their solvating membranes have been reported for various transporters, channels and signal receptors; however, generalizable mechanistic principles governing IMP regulation by lipid environments are neither widely appreciated nor completely understood. Here, we review recent insights into the inter-relationships between complex lipidomes of mammalian membranes, the membrane physicochemical properties resulting from such lipid collectives, and the regulation of IMPs by either or both. The recent proliferation of high-resolution methods to study such lipid-protein interactions has led to generalizable insights, which we synthesize into a general framework termed the 'functional paralipidome' to understand the mutual regulation between membrane proteins and their surrounding lipid microenvironments.
© 2022. Springer Nature Limited.
Figures

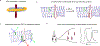
References
-
- Krogh A, Larsson B, von Heijne G & Sonnhammer EL Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of molecular biology 305, 567–580 (2001). - PubMed
-
- Overington JP, Al-Lazikani B & Hopkins AL How many drug targets are there? Nature reviews. Drug discovery 5, 993–996 (2006). - PubMed
-
- Payandeh J & Volgraf M Ligand binding at the protein-lipid interface: strategic considerations for drug design. Nature reviews. Drug discovery 20, 710–722 (2021). - PubMed
-
- Harayama T & Riezman H Understanding the diversity of membrane lipid composition. Nat Rev Mol Cell Biol 19, 281–296 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

