3D proximal tubule-on-chip model derived from kidney organoids with improved drug uptake
- PMID: 36056134
- PMCID: PMC9440090
- DOI: 10.1038/s41598-022-19293-3
3D proximal tubule-on-chip model derived from kidney organoids with improved drug uptake
Abstract
Three-dimensional, organ-on-chip models that recapitulate kidney tissue are needed for drug screening and disease modeling. Here, we report a method for creating a perfusable 3D proximal tubule model composed of epithelial cells isolated from kidney organoids matured under static conditions. These organoid-derived proximal tubule epithelial cells (OPTECs) are seeded in cylindrical channels fully embedded within an extracellular matrix, where they form a confluent monolayer. A second perfusable channel is placed adjacent to each proximal tubule within these reusable multiplexed chips to mimic basolateral drug transport and uptake. Our 3D OPTEC-on-chip model exhibits significant upregulation of organic cation (OCT2) and organic anion (OAT1/3) transporters, which leads to improved drug uptake, compared to control chips based on immortalized proximal tubule epithelial cells. Hence, OPTEC tubules exhibit a higher normalized lactate dehydrogenase (LDH) release, when exposed to known nephrotoxins, cisplatin and aristolochic acid, which are diminished upon adding OCT2 and OAT1/3 transport inhibitors. Our integrated multifluidic platform paves the way for personalized kidney-on-chip models for drug screening and disease modeling.
© 2022. The Author(s).
Conflict of interest statement
Dr. Lewis has been funded by F. Hoffman-La Roche AG, Inc. and AstraZeneca, Inc. Dr. Lewis and Dr. Morizane serve on the Scientific Advisory Board of Trestle Biotherapeutics, which has licensed technology from their labs. The other authors declare no potential conflict of interest.
Figures
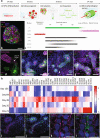


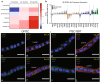
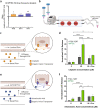
References
-
- Eaton DC, Pooler JP. Vander’s renal physiology. Medicine. 2009;2009:5.
-
- Eric D, Gary M. Drug-induced renal injury. Contin. Med. Educ. 2011;111:349.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

