Tissue-wide metabolomics reveals wide impact of gut microbiota on mice metabolite composition
- PMID: 36056162
- PMCID: PMC9440220
- DOI: 10.1038/s41598-022-19327-w
Tissue-wide metabolomics reveals wide impact of gut microbiota on mice metabolite composition
Abstract
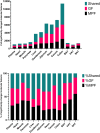
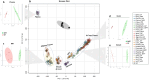
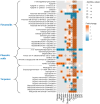
The essential role of gut microbiota in health and disease is well recognized, but the biochemical details that underlie the beneficial impact remain largely undefined. To maintain its stability, microbiota participates in an interactive host-microbiota metabolic signaling, impacting metabolic phenotypes of the host. Dysbiosis of microbiota results in alteration of certain microbial and host metabolites. Identifying these markers could enhance early detection of certain diseases. We report LC-MS based non-targeted metabolic profiling that demonstrates a large effect of gut microbiota on mammalian tissue metabolites. It was hypothesized that gut microbiota influences the overall biochemistry of host metabolome and this effect is tissue-specific. Thirteen different tissues from germ-free (GF) and conventionally-raised (MPF) C57BL/6NTac mice were selected and their metabolic differences were analyzed. Our study demonstrated a large effect of microbiota on mammalian biochemistry at different tissues and resulted in statistically-significant modulation of metabolites from multiple metabolic pathways (p ≤ 0.05). Hundreds of molecular features were detected exclusively in one mouse group, with the majority of these being unique to specific tissue. A vast metabolic response of host to metabolites generated by the microbiota was observed, suggesting gut microbiota has a direct impact on host metabolism.
© 2022. The Author(s).
Conflict of interest statement
Kati Hanhineva (KH), Ville M. Koistinen (VMK), Anton Klåvus (AK), and Ambrin Farizah Babu (AFB) are affiliated with Afekta Technologies Ltd. The remaining authors do not have any competing interests.
Figures






Similar articles
-
Oral Administration of Porphyromonas gingivalis Alters the Gut Microbiome and Serum Metabolome.mSphere. 2018 Oct 17;3(5):e00460-18. doi: 10.1128/mSphere.00460-18. mSphere. 2018. PMID: 30333180 Free PMC article.
-
A meta-analysis study of the robustness and universality of gut microbiome-metabolome associations.Microbiome. 2021 Oct 12;9(1):203. doi: 10.1186/s40168-021-01149-z. Microbiome. 2021. PMID: 34641974 Free PMC article.
-
Gut microbiota influenced the xenograft MC38 tumor growth potentially through interfering host lipid and amino acid metabolisms, basing on the integrated analysis of microbiome and metabolomics.J Chromatogr B Analyt Technol Biomed Life Sci. 2022 Mar 1;1192:123136. doi: 10.1016/j.jchromb.2022.123136. Epub 2022 Feb 5. J Chromatogr B Analyt Technol Biomed Life Sci. 2022. PMID: 35151210
-
Metabolome analysis for investigating host-gut microbiota interactions.J Formos Med Assoc. 2019 Mar;118 Suppl 1:S10-S22. doi: 10.1016/j.jfma.2018.09.007. Epub 2018 Sep 27. J Formos Med Assoc. 2019. PMID: 30269936 Review.
-
Gut Microbiota and Their Associated Metabolites in Diabetes: A Cross Talk Between Host and Microbes-A Review.Metab Syndr Relat Disord. 2023 Feb;21(1):3-15. doi: 10.1089/met.2022.0049. Epub 2022 Oct 26. Metab Syndr Relat Disord. 2023. PMID: 36301254 Review.
Cited by
-
Hypoglycemic Effect of Edible Fungi Polysaccharides Depends on Their Metabolites from the Fermentation of Human Fecal Microbiota.Foods. 2023 Dec 27;13(1):97. doi: 10.3390/foods13010097. Foods. 2023. PMID: 38201125 Free PMC article.
-
Uncovering the core principles of the gut-lung axis to enhance innate immunity in the chicken.Front Immunol. 2022 Oct 4;13:956670. doi: 10.3389/fimmu.2022.956670. eCollection 2022. Front Immunol. 2022. PMID: 36268022 Free PMC article.
-
Eight-year diet and physical activity intervention affects serum metabolites during childhood and adolescence: A nonrandomized controlled trial.iScience. 2024 Jun 17;27(7):110295. doi: 10.1016/j.isci.2024.110295. eCollection 2024 Jul 19. iScience. 2024. PMID: 39055945 Free PMC article.
-
Environmentally Relevant Levels of Antiepileptic Carbamazepine Altered Intestinal Microbial Composition and Metabolites in Amphibian Larvae.Int J Mol Sci. 2024 Jun 25;25(13):6950. doi: 10.3390/ijms25136950. Int J Mol Sci. 2024. PMID: 39000059 Free PMC article.
-
Identification of avaC from Human Gut Microbial Isolates that Converts 5AVA to 2-Piperidone.J Microbiol. 2024 May;62(5):367-379. doi: 10.1007/s12275-024-00141-0. Epub 2024 Jun 17. J Microbiol. 2024. PMID: 38884693 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

