Genomic surveillance: Circulating lineages and genomic variation of SARS-CoV-2 in early pandemic in Ceará state, Northeast Brazil
- PMID: 36057416
- PMCID: PMC9429123
- DOI: 10.1016/j.virusres.2022.198908
Genomic surveillance: Circulating lineages and genomic variation of SARS-CoV-2 in early pandemic in Ceará state, Northeast Brazil
Abstract
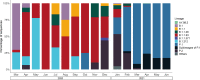
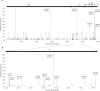
In the Northeast of Brazil, Ceará was the second state most impacted by COVID-19 in number of cases and death rate. Despite that, the early dynamics of the pandemic in Ceará was not yet well understood due the low genomic surveillance of SARS-CoV-2 in 2020. In this study, we analyze the circulating lineages and the genomic variation of the virus in Ceará state. Thirty-four genomes were sequenced and combined with sequences available in GISAID database from March 2020 to June 2021 to compose the study dataset. The most prevalent lineages detected were B.1.1.33, in 2020, and P.1, in 2021. Other lineages were found, such as P.2, sublineages of P.1, B.1, B.1.1, B.1.1.28 and B.1.212. Analyzing the mutations, a total of 202 single-nucleotide polymorphisms (SNPs) were identified among the 34 genomes sequenced, of which 127 were missense, 74 synonymous, and one was a nonsense mutation. Among the missense mutations, C14408T, A23403G, T27299C, G28881A G28883C, and T29148C were the most prevalent within the dataset. Although SARS-CoV-2 sequencing data was limited in 2020, our results could provide insights to better understand the genetic diversity of the circulating lineages in Ceará.
Keywords: COVID-19; Genome sequences; Mutations.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Andrews, S., 2010. A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
-
- Botelho-Souza L.F., Nogueira-Lima F.S., Roca T.P., Naveca F.G., de Oliveria dos Santos A., Maia A.C.S., da Silva C.C., de Melo Mendonça A.L.F., Lugtenburg C.A.B., Azzi C.F.G., Fontes J.L.F., Cavalcante S., de Cássia Pontello Rampazzo R., Santos C.H.N., Di Sabatino Guimarães A.P., Máximo F.R., Villalobos-Salcedo J.M., Vieira D.S. SARS-CoV-2 genomic surveillance in Rondônia, Brazilian Western Amazon. Sci. Rep. 2021;11:1–12. doi: 10.1038/s41598-021-83203-2. - DOI - PMC - PubMed
-
- Candido D.S., Claro I.M., de Jesus J.G., Souza W.M., Moreira F.R.R., Dellicour S., Mellan T.A., du Plessis L., Pereira R.H.M., Sales F.C.S., Manuli E.R., Thézé J., Almeida L., Menezes M.T., Voloch C.M., Fumagalli M.J., Coletti T.M., da Silva C.A.M., Ramundo M.S., Amorim M.R., Hoeltgebaum H.H., Mishra S., Gill M.S., Carvalho L.M., Buss L.F., Prete C.A., Ashworth J., Nakaya H.I., Peixoto P.S., Brady O.J., Nicholls S.M., Tanuri A., Rossi Á.D., Braga C.K.V., Gerber A.L., de Guimarães A.P.C., Gaburo N., Alencar C.S., Ferreira A.C.S., Lima C.X., Levi J.E., Granato C., Ferreira G.M., Francisco R.S., Granja F., Garcia M.T., Moretti M.L., Perroud M.W., Castiñeiras T.M.P.P., Lazari C.S., Hill S.C., de Souza Santos A.A., Simeoni C.L., Forato J., Sposito A.C., Schreiber A.Z., Santos M.N.N., de Sá C.Z., Souza R.P., Resende-Moreira L.C., Teixeira M.M., Hubner J., Leme P.A.F., Moreira R.G., Nogueira M.L., Ferguson N.M., Costa S.F., Proenca-Modena J.L., Vasconcelos A.T.R., Bhatt S., Lemey P., Wu C.H., Rambaut A., Loman N.J., Aguiar R.S., Pybus O.G., Sabino E.C., Faria N.R. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369:1255–1260. doi: 10.1126/science.abd2161. 80-. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

