Patient-Derived Organoids from Colorectal Cancer with Paired Liver Metastasis Reveal Tumor Heterogeneity and Predict Response to Chemotherapy
- PMID: 36058001
- PMCID: PMC9631073
- DOI: 10.1002/advs.202204097
Patient-Derived Organoids from Colorectal Cancer with Paired Liver Metastasis Reveal Tumor Heterogeneity and Predict Response to Chemotherapy
Abstract
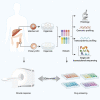
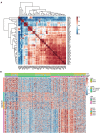
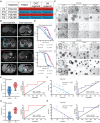
There is no effective method to predict chemotherapy response and postoperative prognosis of colorectal cancer liver metastasis (CRLM) patients. Patient-derived organoid (PDO) has become an important preclinical model. Herein, a living biobank with 50 CRLM organoids derived from primary tumors and paired liver metastatic lesions is successfully constructed. CRLM PDOs from the multiomics levels (histopathology, genome, transcriptome and single-cell sequencing) are comprehensively analyzed and confirmed that this organoid platform for CRLM could capture intra- and interpatient heterogeneity. The chemosensitivity data in vitro reveal the potential value of clinical application for PDOs to predict chemotherapy response (FOLFOX or FOLFIRI) and clinical prognosis of CRLM patients. Taken together, CRLM PDOs can be utilized to deliver a potential application for personalized medicine.
Keywords: chemotherapy response; colorectal cancer liver metastasis; patient-derived organoid; prognosis prediction; tumor heterogeneity.
© 2022 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
H.G.Q. and H.C. are scientific founders of D1 Medical Technology. H.C. is an inventor on several patents related to organoid technology. The remaining authors declare no competing interests with relevance to this study.
Figures




References
-
- Siegel R. L., Miller K. D., Fuchs H. E., Jemal A., Ca‐Cancer J. Clin. 2021, 71, 7. - PubMed
-
- Ren L., Zhu D., A. B. Benson, 3rd , Nordlinger B., Koehne C. H., Delaney C. P., Kerr D., Lenz H. J., Fan J., Wang J., Gu J., Li J., Shen L., Tsarkov P., Tejpar S., Zheng S., Zhang S., Gruenberger T., Qin X., Wang X., Zhang Z., Poston G. J., Xu J., Eur. J. Surg. Oncol. 2020, 46, 955. - PubMed
-
- Power D. G., Kemeny N. E., J. Clin. Oncol. 2010, 28, 2300. - PubMed
-
- Al Bandar M. H., Kim N. K., Oncol. Rep. 2017, 37, 2553. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 82173461/National Natural Science Foundation of China
- U1932145/National Natural Science Foundation of China
- 81972244/National Natural Science Foundation of China
- 82002946/National Natural Science Foundation of China
- YJZZ201802/Fudan University Shanghai Cancer Center Basic and Clinical Translational Research Seed Foundation
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
