Immunosuppressant exposure confounds gene expression analysis in systemic lupus erythematosus
- PMID: 36059457
- PMCID: PMC9430375
- DOI: 10.3389/fimmu.2022.964263
Immunosuppressant exposure confounds gene expression analysis in systemic lupus erythematosus
Abstract
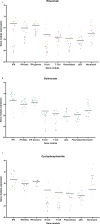
Objectives: The analysis of gene module expression in SLE is emerging as a tool to identify active biological pathways, with the aim of developing targeted therapies for subsets of patients. Detailed information on the effect of immunosuppressants on gene module expression is lacking. We aimed to examine the impact of medication exposure on gene module expression.
Methods: A set of commercially available disease-relevant gene modules were measured in 730 whole blood samples from a dedicated lupus clinic on whom prospectively collected, contemporaneous clinical data including medication exposure were available.
Results: Compared to heathy controls, SLE patients showed over-expression of IFN and under-expression of B cell, T cell and pDC modules. Neutrophil module over-expression and under-expression of B and T cell modules were observed in patients with active lupus nephritis or highly active disease (SLEDAI-2K > 8), while Lupus Low Disease Activity State (LLDAS) had inverse associations. Disease activity in other organ domains was not associated with specific gene modules. In contrast, medications were associated with multiple effects. Glucocorticoid use was associated with under-expression of T cell, B cell and plasmablast modules, and over-expression of neutrophil modules. Mycophenolate and azathioprine exposure were associated with plasmablast module and B cell module under-expression respectively. Disease activity associations with neutrophil over-expression and lymphocyte module under-expression were attenuated by multivariable adjustment for medication exposure.
Conclusion: Medications have significant effect on gene module expression in SLE patients. These findings emphasize the need to control for medications in studies of gene expression in SLE.
Keywords: gene modules; glucocorticoids; immunosuppressants; systemic lupus erythematosus; transcriptional profiling.
Copyright © 2022 Northcott, Gearing, Bonin, Koelmeyer, Hoi, Hertzog and Morand.
Conflict of interest statement
Outside the scope of this work, EM has received consulting fees from AbbVie, AstraZeneca, Amgen, Biogen, Bristol Myers Squibb, Eli Lilly, EMD Serono, Genetech, Janssen, Servier and UCB, and research grants from AbbVie, AstraZeneca, Bristol Myers Squibb, Eli Lilly, EMD Serono and Janssen. AH has received research grants from AstraZeneca, Merck and GSK, and consulting fees from Abbvie, AstraZeneca, BMS, GSK, Janssen and Pfizer. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

