Centaur antibodies: Engineered chimeric equine-human recombinant antibodies
- PMID: 36059507
- PMCID: PMC9437483
- DOI: 10.3389/fimmu.2022.942317
Centaur antibodies: Engineered chimeric equine-human recombinant antibodies
Abstract
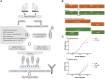
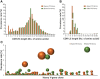
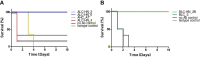
Hyper-immune antisera from large mammals, in particular horses, are routinely used for life-saving anti-intoxication intervention. While highly efficient, the use of these immunotherapeutics is complicated by possible recipient reactogenicity and limited availability. Accordingly, there is an urgent need for alternative improved next-generation immunotherapies to respond to this issue of high public health priority. Here, we document the development of previously unavailable tools for equine antibody engineering. A novel primer set, EquPD v2020, based on equine V-gene data, was designed for efficient and accurate amplification of rearranged horse antibody V-segments. The primer set served for generation of immune phage display libraries, representing highly diverse V-gene repertoires of horses immunized against botulinum A or B neurotoxins. Highly specific scFv clones were selected and expressed as full-length antibodies, carrying equine V-genes and human Gamma1/Lambda constant genes, to be referred as "Centaur antibodies". Preliminary assessment in a murine model of botulism established their therapeutic potential. The experimental approach detailed in the current report, represents a valuable tool for isolation and engineering of therapeutic equine antibodies.
Keywords: anti-toxins; anti-venoms; antibody engineering; botulinum; equine V-genes; phage display.
Copyright © 2022 Rosenfeld, Alcalay, Zvi, Ben-David, Noy-Porat, Chitlaru, Epstein, Israeli, Lazar, Caspi, Barnea, Dor, Chomsky, Pitel, Makdasi, Zichel and Mazor.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

