Aberrant DNA methylation in multiple myeloma: A major obstacle or an opportunity?
- PMID: 36059621
- PMCID: PMC9434119
- DOI: 10.3389/fonc.2022.979569
Aberrant DNA methylation in multiple myeloma: A major obstacle or an opportunity?
Abstract
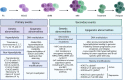
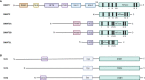
Drug resistance (DR) of cancer cells leading to relapse is a huge problem nowadays to achieve long-lasting cures for cancer patients. This also holds true for the incurable hematological malignancy multiple myeloma (MM), which is characterized by the accumulation of malignant plasma cells in the bone marrow (BM). Although new treatment approaches combining immunomodulatory drugs, corticosteroids, proteasome inhibitors, alkylating agents, and monoclonal antibodies have significantly improved median life expectancy, MM remains incurable due to the development of DR, with the underlying mechanisms remaining largely ill-defined. It is well-known that MM is a heterogeneous disease, encompassing both genetic and epigenetic aberrations. In normal circumstances, epigenetic modifications, including DNA methylation and posttranslational histone modifications, play an important role in proper chromatin structure and transcriptional regulation. However, in MM, numerous epigenetic defects or so-called 'epimutations' have been observed and this especially at the level of DNA methylation. These include genome-wide DNA hypomethylation, locus specific hypermethylation and somatic mutations, copy number variations and/or deregulated expression patterns in DNA methylation modifiers and regulators. The aberrant DNA methylation patterns lead to reduced gene expression of tumor suppressor genes, genomic instability, DR, disease progression, and high-risk disease. In addition, the frequency of somatic mutations in the DNA methylation modifiers seems increased in relapsed patients, again suggesting a role in DR and relapse. In this review, we discuss the recent advances in understanding the involvement of aberrant DNA methylation patterns and/or DNA methylation modifiers in MM development, progression, and relapse. In addition, we discuss their involvement in MM cell plasticity, driving myeloma cells to a cancer stem cell state characterized by a more immature and drug-resistant phenotype. Finally, we briefly touch upon the potential of DNA methyltransferase inhibitors to prevent relapse after treatment with the current standard of care agents and/or new, promising (immuno) therapies.
Keywords: DNA methylation modifiers; DNMTi; MM cell plasticity; epigenetics; multiple myeloma.
Copyright © 2022 Muylaert, Van Hemelrijck, Maes, De Veirman, Menu, Vanderkerken and De Bruyne.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
The Epigenome in Multiple Myeloma: Impact on Tumor Cell Plasticity and Drug Response.Front Oncol. 2018 Dec 11;8:566. doi: 10.3389/fonc.2018.00566. eCollection 2018. Front Oncol. 2018. PMID: 30619733 Free PMC article. Review.
-
Role of epigenetics-microRNA axis in drug resistance of multiple myeloma.J Hematol Oncol. 2017 Jun 17;10(1):121. doi: 10.1186/s13045-017-0492-1. J Hematol Oncol. 2017. PMID: 28623912 Free PMC article. Review.
-
The Role of Epigenetics in the Development and Progression of Multiple Myeloma.Biomedicines. 2022 Oct 31;10(11):2767. doi: 10.3390/biomedicines10112767. Biomedicines. 2022. PMID: 36359286 Free PMC article. Review.
-
Epigenetics in multiple myeloma: From mechanisms to therapy.Semin Cancer Biol. 2018 Aug;51:101-115. doi: 10.1016/j.semcancer.2017.09.007. Epub 2017 Sep 27. Semin Cancer Biol. 2018. PMID: 28962927 Review.
-
Integrative analysis of DNA copy number, DNA methylation and gene expression in multiple myeloma reveals alterations related to relapse.Oncotarget. 2016 Dec 6;7(49):80664-80679. doi: 10.18632/oncotarget.13025. Oncotarget. 2016. PMID: 27811368 Free PMC article.
Cited by
-
Therapeutic potential of natural antisense transcripts and various mechanisms involved for clinical applications and disease prevention.RNA Biol. 2024 Jan;21(1):1-18. doi: 10.1080/15476286.2023.2293335. Epub 2023 Dec 13. RNA Biol. 2024. PMID: 38090817 Free PMC article. Review.
-
The de novo DNA methyltransferase 3B is a novel epigenetic regulator of MYC in multiple myeloma, representing a promising therapeutic target to counter relapse.J Exp Clin Cancer Res. 2025 Apr 17;44(1):125. doi: 10.1186/s13046-025-03382-y. J Exp Clin Cancer Res. 2025. PMID: 40241199 Free PMC article.
-
m1A regulator‑mediated methylation modifications and gene signatures and their prognostic value in multiple myeloma.Exp Ther Med. 2024 Nov 18;29(1):18. doi: 10.3892/etm.2024.12768. eCollection 2025 Jan. Exp Ther Med. 2024. PMID: 39624591 Free PMC article.
-
The Genetic and Molecular Drivers of Multiple Myeloma: Current Insights, Clinical Implications, and the Path Forward.Pharmgenomics Pers Med. 2024 Dec 21;17:573-609. doi: 10.2147/PGPM.S350238. eCollection 2024. Pharmgenomics Pers Med. 2024. PMID: 39723112 Free PMC article. Review.
-
DNA methylation in circulating leukocytes is a novel biomarker in multiple myeloma.Bone Marrow Transplant. 2023 Mar;58(3):334-336. doi: 10.1038/s41409-022-01887-0. Epub 2022 Dec 2. Bone Marrow Transplant. 2023. PMID: 36460820 No abstract available.
References
Publication types
LinkOut - more resources
Full Text Sources

