Efficient and accurate diagnosis of otomycosis using an ensemble deep-learning model
- PMID: 36060244
- PMCID: PMC9437247
- DOI: 10.3389/fmolb.2022.951432
Efficient and accurate diagnosis of otomycosis using an ensemble deep-learning model
Abstract
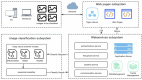
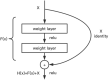
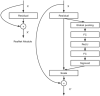
Otomycosis accounts for over 15% of cases of external otitis worldwide. It is common in humid regions and Chinese cultures with ear-cleaning custom. Aspergillus and Candida are the major pathogens causing long-term infection. Early endoscopic and microbiological examinations, performed by otologists and microbiologists, respectively, are important for the appropriate medical treatment of otomycosis. The deep-learning model is a novel automatic diagnostic program that provides quick and accurate diagnoses using a large database of images acquired in clinical settings. The aim of the present study was to introduce a machine-learning model to accurately and quickly diagnose otomycosis caused by Aspergillus and Candida. We propose a computer-aided decision-making system based on a deep-learning model comprising two subsystems: Java web application and image classification. The web application subsystem provides a user-friendly webpage to collect consulted images and display the calculation results. The image classification subsystem mainly trained neural network models for end-to-end data inference. The end user uploads a few images obtained with the ear endoscope, and the system returns the classification results to the user in the form of category probability values. To accurately diagnose otomycosis, we used otoendoscopic images and fungal culture secretion. Fungal fluorescence, culture, and DNA sequencing were performed to confirm the pathogens Aspergillus or Candida spp. In addition, impacted cerumen, external otitis, and normal external auditory canal endoscopic images were retained for reference. We merged these four types of images into an otoendoscopic image gallery. To achieve better accuracy and generalization abilities after model-training, we selected 2,182 of approximately 4,000 ear endoscopic images as training samples and 475 as validation samples. After selecting the deep neural network models, we tested the ResNet, SENet, and EfficientNet neural network models with different numbers of layers. Considering the accuracy and operation speed, we finally chose the EfficientNetB6 model, and the probability values of the four categories of otomycosis, impacted cerumen, external otitis, and normal cases were outputted. After multiple model training iterations, the average accuracy of the overall validation sample reached 92.42%. The results suggest that the system could be used as a reference for general practitioners to obtain more accurate diagnoses of otomycosis.
Keywords: Aspergillus; Candida; deep-learning; otoendoscopic; otomycosis.
Copyright © 2022 Mao, Li, Hu, Wang, Peng, Wang and Sun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Cai Y., Yu J. G., Chen Y., Liu C., Xiao L., Grais M. E., et al. (2021). Investigating the use of a two-stage attention-aware convolutional neural network for the automated diagnosis of otitis media from tympanic membrane images: a prediction model development and validation study. BMJ Open 11, e041139. 10.1136/bmjopen-2020-041139 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

