5-methylcytosine turnover: Mechanisms and therapeutic implications in cancer
- PMID: 36060265
- PMCID: PMC9428128
- DOI: 10.3389/fmolb.2022.976862
5-methylcytosine turnover: Mechanisms and therapeutic implications in cancer
Abstract
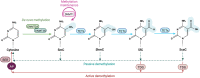
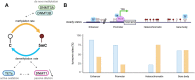
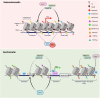
DNA methylation at the fifth position of cytosine (5mC) is one of the most studied epigenetic mechanisms essential for the control of gene expression and for many other biological processes including genomic imprinting, X chromosome inactivation and genome stability. Over the last years, accumulating evidence suggest that DNA methylation is a highly dynamic mechanism driven by a balance between methylation by DNMTs and TET-mediated demethylation processes. However, one of the main challenges is to understand the dynamics underlying steady state DNA methylation levels. In this review article, we give an overview of the latest advances highlighting DNA methylation as a dynamic cycling process with a continuous turnover of cytosine modifications. We describe the cooperative actions of DNMT and TET enzymes which combine with many additional parameters including chromatin environment and protein partners to govern 5mC turnover. We also discuss how mathematical models can be used to address variable methylation levels during development and explain cell-type epigenetic heterogeneity locally but also at the genome scale. Finally, we review the therapeutic implications of these discoveries with the use of both epigenetic clocks as predictors and the development of epidrugs that target the DNA methylation/demethylation machinery. Together, these discoveries unveil with unprecedented detail how dynamic is DNA methylation during development, underlying the establishment of heterogeneous DNA methylation landscapes which could be altered in aging, diseases and cancer.
Keywords: 5-Methylcytosine; DNA methylation; DNMT; TET; cancer; dynamics.
Copyright © 2022 Turpin and Salbert.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Achour M., Mousli M., Alhosin M., Ibrahim A., Peluso J., Muller C. D., et al. (2013). Epigallocatechin-3-gallate up-regulates tumor suppressor gene expression via a reactive oxygen species-dependent down-regulation of UHRF1. Biochem. Biophys. Res. Commun. 430, 208–212. 10.1016/j.bbrc.2012.11.087 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

