Identification and Validation of Immune Markers in Coronary Heart Disease
- PMID: 36060667
- PMCID: PMC9439891
- DOI: 10.1155/2022/2877679
Identification and Validation of Immune Markers in Coronary Heart Disease
Retraction in
-
Retracted: Identification and Validation of Immune Markers in Coronary Heart Disease.Comput Math Methods Med. 2023 Nov 29;2023:9763590. doi: 10.1155/2023/9763590. eCollection 2023. Comput Math Methods Med. 2023. PMID: 38077869 Free PMC article.
Abstract
Background: Coronary heart disease (CHD) is an ischemic heart disease involving a variety of immune factors. This study was aimed at investigating unique immune and m6A patterns in patients with CHD by gene expression in peripheral blood mononuclear cells (PBMCs) and at identifying novel immune biomarkers.
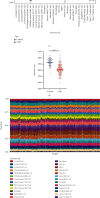
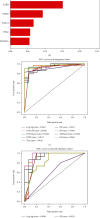
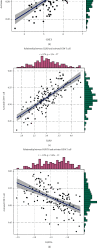
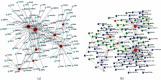
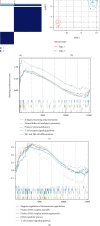
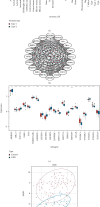
Methods: The CIBERSORT algorithm and single-sample gene set enrichment analysis (ssGSEA) were applied to assess the population of specific infiltrating immunocytes. Weighted Gene Coexpression Network Analysis (WGCNA) was utilized on immune genes matching CHD. A prediction model based on core immune genes was constructed and verified by a machine learning model. Unsupervised cluster analysis identified various immune patterns in the CHD group according to the abundance of immune cells. Methylation of N6 adenosine- (m6A-) related gene was identified from the literature, and t-distributed stochastic neighbor embedding (t-SNE) analysis was used to determine the rationality of the m6A classification. The association between m6A-related genes and various immune cells was estimated using heat maps.
Results: 22/28 immune-associated cells differed between the CHD and normal groups, and a significant difference was detected in the expression of 21 m6A-related genes. The proportion of immune-related cells (activated CD4+ T cells and CD8+ T cells) in the peripheral blood of the CHD group was lower than that of the normal group. The immune genes were divided into four modules, of which the turquoise modules showed a significant association with coronary heart disease. Eight hub immune genes (PDGFRA, GNLY, OSMR, NUDT6, FGFR2, IL2RB, TPM2, and S100A1) can well distinguish the CHD group from the normal group. Two different immune patterns were identified in the CHD group. Interestingly, a significant association was detected between the m6A-related genes and immune cell abundance.
Conclusion: In conclusion, we identified different immune and m6A patterns in CHD. Thus, it could be speculated that the immune system plays a crucial role in CHD, and m6A is correlated with immune genes.
Copyright © 2022 Yuxiong Pan et al.
Conflict of interest statement
The authors have declared that no competing interest exists.
Figures

References
-
- Kott K. A., Vernon S. T., Hansen T., et al. Single-cell immune profiling in coronary artery disease: the role of state-of-the-art immunophenotyping with mass cytometry in the diagnosis of atherosclerosis. Journal of the American Heart Association . 2020;9(24, article e017759) doi: 10.1161/JAHA.120.017759. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

