Proteasomal subunit depletions differentially affect germline integrity in C. elegans
- PMID: 36060813
- PMCID: PMC9428126
- DOI: 10.3389/fcell.2022.901320
Proteasomal subunit depletions differentially affect germline integrity in C. elegans
Abstract
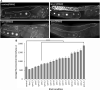
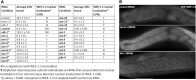
The 26S proteasome is a multi-subunit protein complex that is canonically known for its ability to degrade proteins in cells and maintain protein homeostasis. Non-canonical or non-proteolytic roles of proteasomal subunits exist but remain less well studied. We provide characterization of germline-specific functions of different 19S proteasome regulatory particle (RP) subunits in C. elegans using RNAi specifically from the L4 stage and through generation of endogenously tagged 19S RP lid subunit strains. We show functions for the 19S RP in regulation of proliferation and maintenance of integrity of mitotic zone nuclei, in polymerization of the synaptonemal complex (SC) onto meiotic chromosomes and in the timing of SC subunit redistribution to the short arm of the bivalent, and in turnover of XND-1 proteins at late pachytene. Furthermore, we report that certain 19S RP subunits are required for proper germ line localization of WEE-1.3, a major meiotic kinase. Additionally, endogenous fluorescent labeling revealed that the two isoforms of the essential 19S RP proteasome subunit RPN-6.1 are expressed in a tissue-specific manner in the hermaphrodite. Also, we demonstrate that the 19S RP subunits RPN-6.1 and RPN-7 are crucial for the nuclear localization of the lid subunits RPN-8 and RPN-9 in oocytes, further supporting the ability to utilize the C. elegans germ line as a model to study proteasome assembly real-time. Collectively, our data support the premise that certain 19S RP proteasome subunits are playing tissue-specific roles, especially in the germ line. We propose C. elegans as a versatile multicellular model to study the diverse proteolytic and non-proteolytic roles that proteasome subunits play in vivo.
Keywords: 19S regulatory particle; C. elegans; germ line; meiosis; proteasome.
Copyright © 2022 Fernando, Quesada-Candela, Murray, Ugoaru, Yanowitz and Allen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Adams J., Palombella V. J., Sausville E. A., Johnson J., Destree A., Lazarus D. D., et al. (1999). Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res. 59 (11), 2615–2622. - PubMed
LinkOut - more resources
Full Text Sources

