Comprehensive Analysis of N6-Methyladenosine RNA Methylation Regulators in the Diagnosis and Subtype Classification of Acute Myocardial Infarction
- PMID: 36061306
- PMCID: PMC9433256
- DOI: 10.1155/2022/5173761
Comprehensive Analysis of N6-Methyladenosine RNA Methylation Regulators in the Diagnosis and Subtype Classification of Acute Myocardial Infarction
Abstract
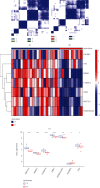
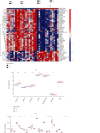
Acute myocardial infarction (AMI) is still a huge danger to human health. Sensitive markers are necessary for the prediction of the risk of AMI and would be beneficial for managing the incidence rate. N6-methyladenosine (m6A) RNA methylation regulators have been confirmed to be involved in the development of various diseases. However, their function in AMI has not been fully elucidated. The purpose of this study was to determine the expression of m6A RNA methylation regulators in AMI as well as their possible functions and prognostic values. The GEO database was used to get the gene expression profiles of patients with and without AMI, and bioinformatics assays of genes with differently expressed expression were performed. We establish two separate m6A subtypes, and relationships between subtypes and immunity were studied. In this study, we identified IGF2BP1, FTO, RBM15, METTL3, YTHDC2, FMR1, and HNRNPA2B1 as the seven major m6A regulators. A nomogram model was developed and confirmed. The consensus clustering algorithm was conducted to categorize AMI patients into two m6A subtypes from the identified m6A regulators. Patients who have activated T-cell activities were found to be in clusterA; they may have a better prognosis as a result. Importantly, we found that patients with high METTL3 expressions had an increased level of Activated.CD4.T.cell and Type.2.T.helper.cell, while having a decreased level of CD56bright.natural.killer.cell, Macrophage, Monocyte, Natural.killer.cell, and Type.17.T.helper.cell. Overall, a diagnostic model of AMI was established based on the genes of IGF2BP1, FTO, RBM15, METTL3, YTHDC2, FMR1, and HNRNPA2B1. Our investigation of m6A subtypes may prove useful in the developments of therapy approaches for AMI.
Copyright © 2022 Xianpei Wang et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

