Transcriptomic analysis of CO2-treated strawberries (Fragaria vesca) with enhanced resistance to softening and oxidative stress at consumption
- PMID: 36061763
- PMCID: PMC9437593
- DOI: 10.3389/fpls.2022.983976
Transcriptomic analysis of CO2-treated strawberries (Fragaria vesca) with enhanced resistance to softening and oxidative stress at consumption
Abstract
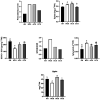
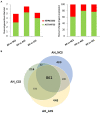
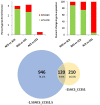
One of the greatest threats to wild strawberries (Fragaria vesca Mara des Bois) after harvest is the highly perishability at ambient temperature. Breeders have successfully met the quality demands of consumers, but the prevention of waste after harvest in fleshy fruits is still pending. Most of the waste is due to the accelerated progress of senescence-like process after harvest linked to a rapid loss of water and firmness at ambient temperature. The storage life of strawberries increases at low temperature, but their quality is limited by the loss of cell structure. The application of high CO2 concentrations increased firmness during cold storage. However, the key genes related to resistance to softening and cell wall disassembly following transference from cold storage at 20°C remain unclear. Therefore, we performed RNA-seq analysis, constructing a weighted gene co-expression network analysis (WGCNA) to identify which molecular determinants play a role in cell wall integrity, using strawberries with contrasting storage conditions, CO2-cold stored (CCS), air-cold stored (ACS), non-cold stored (NCS) kept at ambient temperature, and intact fruit at harvest (AH). The hub genes associated with the cell wall structural architecture of firmer CO2-treated strawberries revealed xyloglucans stabilization attributed mainly to a down-regulation of Csl E1, XTH 15, Exp-like B1 and the maintenance of expression levels of nucleotide sugars transferases such as GMP and FUT as well as improved lamella integrity linked to a down-regulation of RG-lyase, PL-like and PME. The preservation of cell wall elasticity together with the up-regulation of LEA, EXPA4, and MATE, required to maintain cell turgor, is the mechanisms controlled by high CO2. In stressed air-cold stored strawberries, in addition to an acute softening, there is a preferential transcript accumulation of genes involved in lignin and raffinose pathways. Non-cold stored strawberries kept at 20°C after harvest are characterized by an enrichment in genes mainly involved in oxidative stress and up-expression of genes involved in jasmonate biosynthesis. The present results on transcriptomic analysis of CO2-treated strawberries with enhanced resistance to softening and oxidative stress at consumption will help to improve breeding strategies of both wild and cultivated strawberries.
Keywords: CO2; H2O2; RNA-seq; cell wall; firmness; lignin; strawberries; xyloglucans.
Copyright © 2022 del Olmo, Romero, Alvarez, Tarradas, Sanchez-Ballesta, Escribano and Merodio.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures









References
-
- Bang J., Lim S., Yi G., Lee J. G., Lee E. J. (2019). Integrated transcriptomic-metabolomic analysis reveals cellular responses of harvested strawberry fruit subjected to short-term exposure to high levels of carbon dioxide. Postharvest Biol. Technol. 148, 120–131. doi: 10.1016/j.postharvbio.2018.11.003 - DOI
-
- Blanch M., Álvarez I., Sanchez-Ballesta M. T., Escribano M. I., Merodio C. (2019). Involvement of fatty acids in the response to high CO2 and low temperature in harvested strawberries. Postharvest Biol. Technol. 147, 196–205. doi: 10.1016/j.postharvbio.2018.10.001 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

