Overexpression of leucoanthocyanidin reductase or anthocyanidin reductase elevates tannins content and confers cassava resistance to two-spotted spider mite
- PMID: 36061805
- PMCID: PMC9433999
- DOI: 10.3389/fpls.2022.994866
Overexpression of leucoanthocyanidin reductase or anthocyanidin reductase elevates tannins content and confers cassava resistance to two-spotted spider mite
Abstract
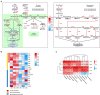
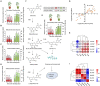
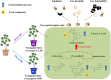
The two-spotted spider mite (TSSM) is a destructive cassava pest. Intensive demonstration of resistance mechanism greatly facilitates the creation of TSSM-resistant cassava germplasm. Gene to metabolite network plays a crucial role in modulating plant resistance, but little is known about the genes and related metabolites which are responsible for cassava resistance to TSSM. Here, a highly resistant (HR) and a highly susceptible (HS) cassava cultivar were used, integrative and comparative transcriptomic and metabolomic analyses between these two cultivars after TSSM infestation revealed that several genes and metabolites were closely related and significantly different in abundance. In particular, the expression of leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR) genes showed a high positive correlation with most of the metabolites in the tannin biosynthesis pathway. Furthermore, transgenic cassava lines overexpressing either of the genes elevated tannin concentrations and conferred cassava resistance to TSSM. Additionally, different forms of tannins possessed distinct bioactivity on TSSM, of which total condensed tannins (LC50 = 375.68 mg/l) showed maximum lethal effects followed by procyanidin B1 (LC50 = 3537.10 mg/l). This study accurately targets LAR, ANR and specific tannin compounds as critical genes and metabolites in shaping cassava resistance to TSSM, which could be considered as biomarkers for evaluation and creation of pest-resistant cassava germplasm.
Keywords: anthocyanidin reductase; cassava (Manihot esculenta Crantz); leucoanthocyanidin reductase; resistance mechanism; tannins (condensed); two-spotted spider mite (Tetranychus urticae Koch).
Copyright © 2022 Chen, Liang, Wu, Liu, Liu, Zhao, Li, Chen, Wang, Han, Wu, Yao, Shui, Qiao, Zhan and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
MePAL6 regulates lignin accumulation to shape cassava resistance against two-spotted spider mite.Front Plant Sci. 2023 Jan 6;13:1067695. doi: 10.3389/fpls.2022.1067695. eCollection 2022. Front Plant Sci. 2023. PMID: 36684737 Free PMC article.
-
Identification of cassava germplasms resistant to two-spotted spider mite in China: From greenhouse large-scale screening to field validation.Front Plant Sci. 2022 Dec 7;13:1054909. doi: 10.3389/fpls.2022.1054909. eCollection 2022. Front Plant Sci. 2022. PMID: 36570903 Free PMC article.
-
Exogenous methyl jasmonate induced cassava defense response and enhanced resistance to Tetranychus urticae.Exp Appl Acarol. 2023 Jan;89(1):45-60. doi: 10.1007/s10493-022-00773-0. Epub 2023 Jan 12. Exp Appl Acarol. 2023. PMID: 36635606
-
Plant-Herbivore Interactions: A Case of an Extreme Generalist, the Two-Spotted Spider Mite Tetranychus urticae.Mol Plant Microbe Interact. 2017 Dec;30(12):935-945. doi: 10.1094/MPMI-07-17-0168-CR. Epub 2017 Oct 23. Mol Plant Microbe Interact. 2017. PMID: 28857675 Review.
-
Current knowledge and future research perspectives on cassava (Manihot esculenta Crantz) chemical defenses: An agroecological view.Phytochemistry. 2016 Oct;130:10-21. doi: 10.1016/j.phytochem.2016.05.013. Epub 2016 Jun 14. Phytochemistry. 2016. PMID: 27316676 Review.
Cited by
-
MePAL6 regulates lignin accumulation to shape cassava resistance against two-spotted spider mite.Front Plant Sci. 2023 Jan 6;13:1067695. doi: 10.3389/fpls.2022.1067695. eCollection 2022. Front Plant Sci. 2023. PMID: 36684737 Free PMC article.
-
Trichomes and unique gene expression confer insect herbivory resistance in Vitis labrusca grapevines.BMC Plant Biol. 2024 Jun 27;24(1):609. doi: 10.1186/s12870-024-05260-9. BMC Plant Biol. 2024. PMID: 38926877 Free PMC article.
-
ABA and MeJA Induced Catechin and Epicatechin Biosynthesis and Accumulation in Camellia oleifera Fruit Shells.Plants (Basel). 2024 Aug 9;13(16):2211. doi: 10.3390/plants13162211. Plants (Basel). 2024. PMID: 39204647 Free PMC article.
-
Overexpression of LAR1 Suppresses Anthocyanin Biosynthesis by Enhancing Catechin Competition Leading to Promotion of Proanthocyanidin Pathway in Spine Grape (Vitis davidii) Cells.Int J Mol Sci. 2024 Nov 11;25(22):12087. doi: 10.3390/ijms252212087. Int J Mol Sci. 2024. PMID: 39596158 Free PMC article.
-
Advancements and strategies of genetic improvement in cassava (Manihot esculenta Crantz): from conventional to genomic approaches.Hortic Res. 2024 Dec 2;12(3):uhae341. doi: 10.1093/hr/uhae341. eCollection 2025 Mar. Hortic Res. 2024. PMID: 40061801 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

