CRISPR/Cas9 mediated gene-editing of GmHdz4 transcription factor enhances drought tolerance in soybean (Glycine max [L.] Merr.)
- PMID: 36061810
- PMCID: PMC9437544
- DOI: 10.3389/fpls.2022.988505
CRISPR/Cas9 mediated gene-editing of GmHdz4 transcription factor enhances drought tolerance in soybean (Glycine max [L.] Merr.)
Abstract
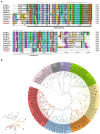
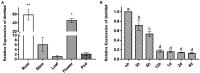
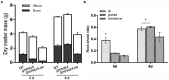
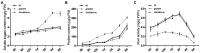
The HD-Zip transcription factors play a crucial role in plant development, secondary metabolism, and abiotic stress responses, but little is known about HD-Zip I genes in soybean. Here, a homeodomain-leucine zipper gene designated GmHdz4 was isolated. Chimeric soybean plants, GmHdz4 overexpressing (GmHdz4-oe), and gene-editing via CRISPR/Cas9 (gmhdz4) in hairy roots, were generated to examine the GmHdz4 gene response to polyethylene glycol (PEG)-simulated drought stress. Bioinformatic analysis showed GmHdz4 belonged to clade δ, and was closely related to other drought tolerance-related HD-Zip I family genes such as AtHB12, Oshox12, and Gshdz4. The GmHdz4 was located in the plant nucleus and showed transcriptional activation activity by yeast hybrid assay. Quantitative real-time PCR analysis revealed that GmHdz4 expression varied in tissues and was induced by PEG-simulated drought stress. The gmhdz4 showed promoted growth of aboveground parts, and its root system architecture, including the total root length, the root superficial area, and the number of root tips were significantly higher than those of GmHdz4-oe even the non-transgenic line (NT) on root tips number. The better maintenance of turgor pressure by osmolyte accumulation, and the higher activity of antioxidant enzymes to scavenge reactive oxygen species, ultimately suppressed the accumulation of hydrogen peroxide (H2O2), superoxide anion (O2-), and malondialdehyde (MDA), conferring higher drought tolerance in gmhdz4 compared with both GmHdz4-oe and NT. Together, our results provide new insights for future research on the mechanisms by which GmHdz4 gene-editing via CRISPR/Cas9 system could promote drought stress and provide a potential target for molecular breeding in soybean.
Keywords: Glycine max; GmHdz4; HD-ZIP; drought stress; root system architecture.
Copyright © 2022 Zhong, Hong, Shu, Li, Liu, Chen, Islam, Zhou and Tang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Alexieva V., Sergiev I., Mapelli S., Karanov E. (2001). The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat. Plant Cell Envi. 24, 1337–1344. doi: 10.1046/j.1365-3040.2001.00778.x - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

