Computational prediction and experimental validation of Salmonella Typhimurium SopE-mediated fine-tuning of autophagy in intestinal epithelial cells
- PMID: 36061866
- PMCID: PMC9428466
- DOI: 10.3389/fcimb.2022.834895
Computational prediction and experimental validation of Salmonella Typhimurium SopE-mediated fine-tuning of autophagy in intestinal epithelial cells
Abstract
Macroautophagy is a ubiquitous homeostasis and health-promoting recycling process of eukaryotic cells, targeting misfolded proteins, damaged organelles and intracellular infectious agents. Some intracellular pathogens such as Salmonella enterica serovar Typhimurium hijack this process during pathogenesis. Here we investigate potential protein-protein interactions between host transcription factors and secreted effector proteins of Salmonella and their effect on host gene transcription. A systems-level analysis identified Salmonella effector proteins that had the potential to affect core autophagy gene regulation. The effect of a SPI-1 effector protein, SopE, that was predicted to interact with regulatory proteins of the autophagy process, was investigated to validate our approach. We then confirmed experimentally that SopE can directly bind to SP1, a host transcription factor, which modulates the expression of the autophagy gene MAP1LC3B. We also revealed that SopE might have a double role in the modulation of autophagy: Following initial increase of MAP1LC3B transcription triggered by Salmonella infection, subsequent decrease in MAP1LC3B transcription at 6h post-infection was SopE-dependent. SopE also played a role in modulation of the autophagy flux machinery, in particular MAP1LC3B and p62 autophagy proteins, depending on the level of autophagy already taking place. Upon typical infection of epithelial cells, the autophagic flux is increased. However, when autophagy was chemically induced prior to infection, SopE dampened the autophagic flux. The same was also observed when most of the intracellular Salmonella cells were not associated with the SCV (strain lacking sifA) regardless of the autophagy induction status before infection. We demonstrated how regulatory network analysis can be used to better characterise the impact of pathogenic effector proteins, in this case, Salmonella. This study complements previous work in which we had demonstrated that specific pathogen effectors can affect the autophagy process through direct interaction with autophagy proteins. Here we show that effector proteins can also influence the upstream regulation of the process. Such interdisciplinary studies can increase our understanding of the infection process and point out targets important in intestinal epithelial cell defense.
Keywords: Host-microbe interactions; MAP1LC3B; Salmonella Typhimurium; SopE; autophagy; network biology.
Copyright © 2022 Demeter, Jacomin, Gul, Lister, Lipscombe, Invernizzi, Branchu, Macaulay, Nezis, Kingsley, Korcsmaros and Hautefort.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

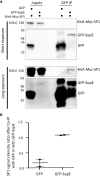

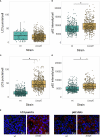

References
-
- Aguilar C., Costa S., Maudet C., Vivek-Ananth R. P., Zaldívar-López S., Garrido J. J., et al. . (2021). Reprogramming of microRNA expression via E2F1 downregulation promotes salmonella infection both in infected and bystander cells. Nat. Commun. 12, 3392. doi: 10.1038/s41467-021-23593-z - DOI - PMC - PubMed
-
- Bakshi C. S., Singh V. P., Wood M. W., Jones P. W., Wallis T. S., Galyov E. E. (2000). Identification of SopE2, a salmonella secreted protein which is highly homologous to SopE and involved in bacterial invasion of epithelial cells. J. Bacteriol. 182, 2341–2344. doi: 10.1128/JB.182.8.2341-2344.2000 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/R012490/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10353/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10355/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/CSP1720/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/T/000PR9819/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/T/000PR9817/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M011216/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/N007964/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M025489/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/R012504/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10348/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10349/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L006324/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P007856/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical

