Multi-type RFC1 repeat expansions as the most common cause of hereditary sensory and autonomic neuropathy
- PMID: 36061987
- PMCID: PMC9428154
- DOI: 10.3389/fneur.2022.986504
Multi-type RFC1 repeat expansions as the most common cause of hereditary sensory and autonomic neuropathy
Abstract
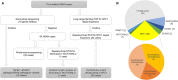
Non-coding repeat expansions within RFC1 and NOTCH2NLC genes have lately been linked to multisystem neurodegenerative diseases, which also shed light on yet undiagnosed patients with inherited peripheral neuropathies. The aim of this study was to identify the genetic basis of patients with hereditary sensory and autonomic neuropathy (HSAN). We collected 79 unrelated DNA samples clinically suspected with HSAN from multiple regions of Japan. Mutation screening was first performed using gene panel sequencing and whole-exome sequencing. Pathogenic/likely pathogenic variants were identified from genes of WNK1/HSN2 (6 cases), SCN9A (3 cases), NTRK1 (3 cases), and DNMT1 (2 cases). Subsequently, long-range flanking PCR and repeat-primed PCR were applied to analyze repeat expansions in RFC1 and NOTCH2NLC. Bi-allelic RFC1 repeat expansions were detected from 20 adult-onset HSAN patients, consisting of [(AAGGG)exp/(AAGGG)exp] (8 cases), [(ACAGG)exp/(ACAGG)exp] (8 cases), and [(AAGGG)exp/(ACAGG)exp] (4 cases). GGC repeat expansion in NOTCH2NLC was found in 1 case. Single-nucleotide variant-based haplotype analysis of patients harboring disease-associated repeat expansions in RFC1 revealed distinguishable haplotypes among subgroups with different repeat genotypes. These findings substantially redefine the genetic spectrum of HSAN, where multi-type RFC1 repeat expansions account for 25.3% of all patients, highlighting the necessity of genetic screening, particularly for adult-onset patients.
Keywords: RFC1; gene panel sequencing; haplotype; non-coding repeat expansion; sensory neuropathy.
Copyright © 2022 Yuan, Higuchi, Ando, Matsuura, Hashiguchi, Yoshimura, Nakamura, Sakiyama, Mitsui, Ishiura, Tsuji and Takashima.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources

