Global phosphoproteomic profiling of skeletal muscle in ovarian hormone-deficient mice
- PMID: 36062884
- PMCID: PMC9639773
- DOI: 10.1152/physiolgenomics.00104.2022
Global phosphoproteomic profiling of skeletal muscle in ovarian hormone-deficient mice
Abstract
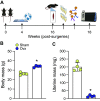
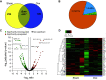
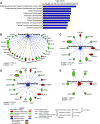
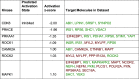
Protein phosphorylation is important in skeletal muscle development, growth, regeneration, and contractile function. Alterations in the skeletal muscle phosphoproteome due to aging have been reported in males; however, studies in females are lacking. We have demonstrated that estrogen deficiency decreases muscle force, which correlates with decreased myosin regulatory light chain phosphorylation. Thus, we questioned whether the decline of estrogen in females that occurs with aging might alter the skeletal muscle phosphoproteome. C57BL/6J female mice (6 mo) were randomly assigned to a sham-operated (Sham) or ovariectomy (Ovx) group to investigate the effects of estrogen deficiency on skeletal muscle protein phosphorylation in a resting, noncontracting condition. After 16 wk of estrogen deficiency, the tibialis anterior muscle was dissected and prepped for label-free nano-liquid chromatography-tandem mass spectrometry phosphoproteomic analysis. We identified 4,780 phosphopeptides in tibialis anterior muscles of ovariectomized (Ovx) and Sham-operated (Sham) control mice. Further analysis revealed 647 differentially regulated phosphopeptides (Benjamini-Hochberg adjusted P value < 0.05 and 1.5-fold change ratio) that corresponded to 130 proteins with 22 proteins differentially phosphorylated (3 unique to Ovx, 2 unique to Sham, 6 upregulated, and 11 downregulated). Differentially phosphorylated proteins associated with the sarcomere, cytoplasm, and metabolic and calcium signaling pathways were identified. Our work provides the first global phosphoproteomic analysis in females and how estrogen deficiency impacts the skeletal muscle phosphoproteome.
Keywords: AMPK; YAP; estrogen; females; ovariectomy.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures



Comment in
-
Emerging roles for estrogen in regulating skeletal muscle physiology.Physiol Genomics. 2023 Feb 1;55(2):75-78. doi: 10.1152/physiolgenomics.00158.2022. Epub 2023 Jan 9. Physiol Genomics. 2023. PMID: 36622080 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

