Systemic proteomics and miRNA profile analysis of exosomes derived from human pluripotent stem cells
- PMID: 36064647
- PMCID: PMC9444124
- DOI: 10.1186/s13287-022-03142-1
Systemic proteomics and miRNA profile analysis of exosomes derived from human pluripotent stem cells
Abstract
Background: Increasing studies have reported the therapeutic effect of mesenchymal stem cell (MSC)-derived exosomes by which protein and miRNA are clearly characterized. However, the proteomics and miRNA profiles of exosomes derived from human embryonic stem cells (hESCs) and human-induced pluripotent stem cells (hiPSCs) remain unclear.
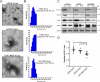
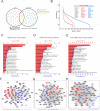
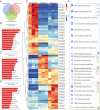
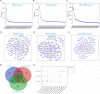
Methods: In this study, we isolated exosomes from hESCs, hiPSCs, and human umbilical cord mesenchymal stem cells (hUC-MSCs) via classic ultracentrifugation and a 0.22-μm filter, followed by the conservative identification. Tandem mass tag labeling and label-free relative peptide quantification together defined their proteomics. High-throughput sequencing was performed to determine miRNA profiles. Then, we conducted a bioinformatics analysis to identify the dominant biological processes and pathways modulated by exosome cargos. Finally, the western blot and RT-qPCR were performed to detect the actual loads of proteins and miRNAs in three types of exosomes.
Results: Based on our study, the cargos from three types of exosomes contribute to sophisticated biological processes. In comparison, hESC exosomes (hESC-Exos) were superior in regulating development, metabolism, and anti-aging, and hiPSC exosomes (hiPSC-Exos) had similar biological functions as hESC-Exos, whereas hUC-MSCs exosomes (hUC-MSC-Exos) contributed more to immune regulation.
Conclusions: The data presented in our study help define the protein and miRNA landscapes of three exosomes, predict their biological functions via systematic and comprehensive network analysis at the system level, and reveal their respective potential applications in different fields so as to optimize exosome selection in preclinical and clinical trials.
Keywords: Exosomes; Human embryonic stem cells; Human umbilical cord mesenchymal stem cells; Human-induced pluripotent stem cells; Proteomics; miRNA.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no potential conflict of interest.
The authors declare no potential conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

