Ectopic JAK-STAT activation enables the transition to a stem-like and multilineage state conferring AR-targeted therapy resistance
- PMID: 36065066
- PMCID: PMC9499870
- DOI: 10.1038/s43018-022-00431-9
Ectopic JAK-STAT activation enables the transition to a stem-like and multilineage state conferring AR-targeted therapy resistance
Erratum in
-
Author Correction: Ectopic JAK-STAT activation enables the transition to a stem-like and multilineage state conferring AR-targeted therapy resistance.Nat Cancer. 2022 Oct;3(10):1271. doi: 10.1038/s43018-022-00458-y. Nat Cancer. 2022. PMID: 36241729 Free PMC article. No abstract available.
Abstract
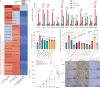
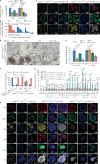
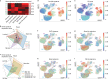
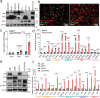
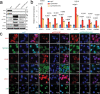
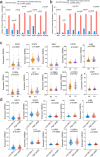
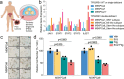
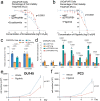
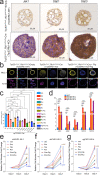
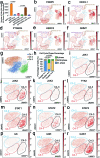
Emerging evidence indicates that various cancers can gain resistance to targeted therapies by acquiring lineage plasticity. Although various genomic and transcriptomic aberrations correlate with lineage plasticity, the molecular mechanisms enabling the acquisition of lineage plasticity have not been fully elucidated. We reveal that Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signaling is a crucial executor in promoting lineage plasticity-driven androgen receptor (AR)-targeted therapy resistance in prostate cancer. Importantly, ectopic JAK-STAT activation is specifically required for the resistance of stem-like subclones expressing multilineage transcriptional programs but not subclones exclusively expressing the neuroendocrine-like lineage program. Both genetic and pharmaceutical inhibition of JAK-STAT signaling resensitizes resistant tumors to AR-targeted therapy. Together, these results suggest that JAK-STAT are compelling therapeutic targets for overcoming lineage plasticity-driven AR-targeted therapy resistance.
© 2022. The Author(s).
Conflict of interest statement
G.R. holds issued and pending patents, which have been licensed to EtiraRx. G.R. serves or has served in an advisory role to Bayer, Johnson and Johnson, Myovant, EtiraRx, Amgen, Pfizer and Astellas. G.R. has or has had grant support from Bayer, EtiraRx and Johnson and Johnson. All other authors declare that they have no competing interests.
Figures








Comment in
-
JAK inhibition shows two faces in prostate cancer.Nat Cancer. 2022 Sep;3(9):1021-1023. doi: 10.1038/s43018-022-00437-3. Nat Cancer. 2022. PMID: 36138136 No abstract available.
References
-
- Bishop JL, et al. The master neural transcription factor BRN2 is an androgen receptor–suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 2017;7:54–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

