Whole-Genome Sequencing Reveals Diversity of Carbapenem-Resistant Pseudomonas aeruginosa Collected through CDC's Emerging Infections Program, United States, 2016-2018
- PMID: 36066241
- PMCID: PMC9487505
- DOI: 10.1128/aac.00496-22
Whole-Genome Sequencing Reveals Diversity of Carbapenem-Resistant Pseudomonas aeruginosa Collected through CDC's Emerging Infections Program, United States, 2016-2018
Abstract
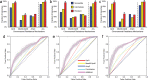
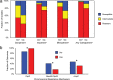
The CDC's Emerging Infections Program (EIP) conducted population- and laboratory-based surveillance of US carbapenem-resistant Pseudomonas aeruginosa (CRPA) from 2016 through 2018. To characterize the pathotype, 1,019 isolates collected through this project underwent antimicrobial susceptibility testing and whole-genome sequencing. Sequenced genomes were classified using the seven-gene multilocus sequence typing (MLST) scheme and a core genome (cg)MLST scheme was used to determine phylogeny. Both chromosomal and horizontally transmitted mechanisms of carbapenem resistance were assessed. There were 336 sequence types (STs) among the 1,019 sequenced genomes, and the genomes varied by an average of 84.7% of the cgMLST alleles used. Mutations associated with dysfunction of the porin OprD were found in 888 (87.1%) of the genomes and were correlated with carbapenem resistance, and a machine learning model incorporating hundreds of genetic variations among the chromosomal mechanisms of resistance was able to classify resistant genomes. While only 7 (0.1%) isolates harbored carbapenemase genes, 66 (6.5%) had acquired non-carbapenemase β-lactamase genes, and these were more likely to have OprD dysfunction and be resistant to all carbapenems tested. The genetic diversity demonstrates that the pathotype includes a variety of strains, and clones previously identified as high-risk make up only a minority of CRPA strains in the United States. The increased carbapenem resistance in isolates with acquired non-carbapenemase β-lactamase genes suggests that horizontally transmitted mechanisms aside from carbapenemases themselves may be important drivers of the spread of carbapenem resistance in P. aeruginosa.
Keywords: Pseudomonas aeruginosa; antibiotic resistance; mechanisms of resistance; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- CDC. 2019. Antibiotic resistance threats in the United States. CDC, Atlanta, GA. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-re....
-
- Nathwani D, Raman G, Sulham K, Gavaghan M, Menon V. 2014. Clinical and economic consequences of hospital-acquired resistant and multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control 3:32. 10.1186/2047-2994-3-32. - DOI - PMC - PubMed
-
- Shortridge D, Gales AC, Streit JM, Huband MD, Tsakris A, Jones RN. 2019. Geographic and temporal patterns of antimicrobial resistance in Pseudomonas aeruginosa over 20 years from the SENTRY antimicrobial surveillance program, 1997–2016. Open Forum Infect Dis 6:S63–S68. 10.1093/ofid/ofy343. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

