A balancing act: interactions within NuA4/TIP60 regulate picNuA4 function in Saccharomyces cerevisiae and humans
- PMID: 36066422
- PMCID: PMC9630986
- DOI: 10.1093/genetics/iyac136
A balancing act: interactions within NuA4/TIP60 regulate picNuA4 function in Saccharomyces cerevisiae and humans
Abstract
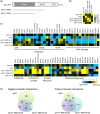
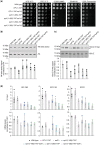
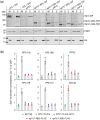
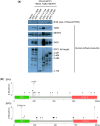
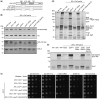
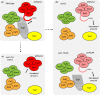
The NuA4 lysine acetyltransferase complex acetylates histone and nonhistone proteins and functions in transcription regulation, cell cycle progression, and DNA repair. NuA4 harbors an interesting duality in that its catalytic module can function independently and distinctly as picNuA4. At the molecular level, picNuA4 anchors to its bigger brother via physical interactions between the C-terminus of Epl1 and the HSA domain of Eaf1, the NuA4 central scaffolding subunit. This is reflected at the regulatory level, as picNuA4 can be liberated genetically from NuA4 by disrupting the Epl1-Eaf1 interaction. As such, removal of either Eaf1 or the Epl1 C-terminus offers a unique opportunity to elucidate the contributions of Eaf1 and Epl1 to NuA4 biology and in turn their roles in balancing picNuA4 and NuA4 activities. Using high-throughput genetic and gene expression profiling, and targeted functional assays to compare eaf1Δ and epl1-CΔ mutants, we found that EAF1 and EPL1 had both overlapping and distinct roles. Strikingly, loss of EAF1 or its HSA domain led to a significant decrease in the amount of picNuA4, while loss of the Epl1 C-terminus increased picNuA4 levels, suggesting starkly opposing effects on picNuA4 regulation. The eaf1Δ epl1-CΔ double mutants resembled the epl1-CΔ single mutants, indicating that Eaf1's role in picNuA4 regulation depended on the Epl1 C-terminus. Key aspects of this regulation were evolutionarily conserved, as truncating an Epl1 homolog in human cells increased the levels of other picNuA4 subunits. Our findings suggested a model in which distinct aspects of the Epl1-Eaf1 interaction regulated picNuA4 amount and activity.
Keywords: chromatin; gene regulation; histone acetylase; histone acetylation; yeast genetics.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures


Similar articles
-
Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin.Genes Dev. 2003 Jun 1;17(11):1415-28. doi: 10.1101/gad.1056603. Genes Dev. 2003. PMID: 12782659 Free PMC article.
-
Structure and flexibility of the yeast NuA4 histone acetyltransferase complex.Elife. 2022 Oct 20;11:e81400. doi: 10.7554/eLife.81400. Elife. 2022. PMID: 36263929 Free PMC article.
-
Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants.Mol Cell Biol. 2008 Apr;28(7):2257-70. doi: 10.1128/MCB.01755-07. Epub 2008 Jan 22. Mol Cell Biol. 2008. PMID: 18212047 Free PMC article.
-
NuA4 and SWR1-C: two chromatin-modifying complexes with overlapping functions and components.Biochem Cell Biol. 2009 Oct;87(5):799-815. doi: 10.1139/O09-062. Biochem Cell Biol. 2009. PMID: 19898529 Review.
-
Reading chromatin: insights from yeast into YEATS domain structure and function.Epigenetics. 2010 Oct 1;5(7):573-7. doi: 10.4161/epi.5.7.12856. Epub 2010 Oct 1. Epigenetics. 2010. PMID: 20657183 Free PMC article. Review.
Cited by
-
Possible regulatory network and associated pathways governing the expression of ADH2 in Saccharomyces cerevisiae.Curr Genet. 2025 Aug 18;71(1):15. doi: 10.1007/s00294-025-01321-0. Curr Genet. 2025. PMID: 40824391 Review.
References
-
- Ausubel M, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. PA: John Wiley & Sons, Inc., Media; 1987.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

