Transcriptome and DNA methylome analyses reveal underlying mechanisms for the racial disparity in uterine fibroids
- PMID: 36066972
- PMCID: PMC9714787
- DOI: 10.1172/jci.insight.160274
Transcriptome and DNA methylome analyses reveal underlying mechanisms for the racial disparity in uterine fibroids
Abstract
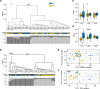
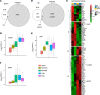
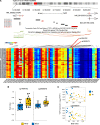
Uterine fibroids (leiomyomas) affect Black women disproportionately compared with women of other races and ethnicities in terms of prevalence, incidence, and severity of symptoms. The causes of this racial disparity are essentially unknown. We hypothesized that myometria of Black women are more susceptible to developing fibroids, and we examined the transcriptomic and DNA methylation profiles of myometria and fibroids from Black and White women for comparison. Myometrial samples cluster by race in both their transcriptome and DNA methylation profiles, whereas fibroid samples only cluster by race in the latter. More differentially expressed genes (DEGs) were detected in the Black and White myometrial sample comparison than in the fibroid comparison. Leiomyoma gene set expression analysis identified 4 clusters of DEGs, including a cluster of 24 genes with higher expression in myometrial samples from Black women. One of the DEGs in this group, von Willibrands factor (VWF), was significantly hypomethylated in both myometrial samples from Black women and in all fibroids at 2 CpG probes that are near a putative enhancer site and that are correlated with VWF expression levels. These results suggest that the molecular basis for the disparity in fibroid disease between Black and White women could be found in the myometria before fibroid development and not in the fibroids themselves.
Keywords: Epigenetics; Molecular genetics; Obstetrics/gynecology; Reproductive Biology.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

