Resident macrophage subpopulations occupy distinct microenvironments in the kidney
- PMID: 36066976
- PMCID: PMC9714795
- DOI: 10.1172/jci.insight.161078
Resident macrophage subpopulations occupy distinct microenvironments in the kidney
Abstract
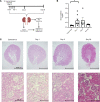
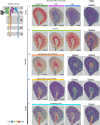
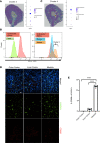
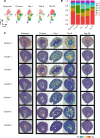
The kidney contains a population of resident macrophages from birth that expands as it grows and forms a contiguous network throughout the tissue. Kidney-resident macrophages (KRMs) are important in homeostasis and the response to acute kidney injury. While the kidney contains many microenvironments, it is unknown whether KRMs are a heterogeneous population differentiated by function and location. We combined single-cell RNA-Seq (scRNA-Seq), spatial transcriptomics, flow cytometry, and immunofluorescence imaging to localize, characterize, and validate KRM populations during quiescence and following 19 minutes of bilateral ischemic kidney injury. scRNA-Seq and spatial transcriptomics revealed 7 distinct KRM subpopulations, which are organized into zones corresponding to regions of the nephron. Each subpopulation was identifiable by a unique transcriptomic signature, suggesting distinct functions. Specific protein markers were identified for 2 clusters, allowing analysis by flow cytometry or immunofluorescence imaging. Following injury, the original localization of each subpopulation was lost, either from changing locations or transcriptomic signatures. The original spatial distribution of KRMs was not fully restored for at least 28 days after injury. The change in KRM localization confirmed a long-hypothesized dysregulation of the local immune system following acute injury and may explain the increased risk for chronic kidney disease.
Keywords: Bioinformatics; Immunology; Innate immunity; Macrophages; Nephrology.
Figures

References
-
- Hume D, Gordon S. Mononuclear phagocyte system of the mouse defined by immunohistochemical localization of antigen F4/80. Identification of resident macrophages in renal medullary and cortical interstitium and the juxtaglomerular complex. J Exp Med. 1983;157(5):1704–1709. doi: 10.1084/jem.157.5.1704. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

