Urban rats as carriers of invasive Salmonella Typhimurium sequence type 313, Kisangani, Democratic Republic of Congo
- PMID: 36067238
- PMCID: PMC9481155
- DOI: 10.1371/journal.pntd.0010740
Urban rats as carriers of invasive Salmonella Typhimurium sequence type 313, Kisangani, Democratic Republic of Congo
Abstract
Background: Invasive non-typhoidal Salmonella (iNTS-mainly serotypes Enteritidis and Typhimurium) are major causes of bloodstream infections in children in sub-Saharan Africa, but their reservoir remains unknown. We assessed iNTS carriage in rats in an urban setting endemic for iNTS carriage and compared genetic profiles of iNTS from rats with those isolated from humans.
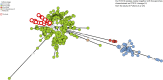
Methodology/principal findings: From April 2016 to December 2018, rats were trapped in five marketplaces and a slaughterhouse in Kisangani, Democratic Republic of the Congo. After euthanasia, blood, liver, spleen, and rectal content were cultured for Salmonella. Genetic relatedness between iNTS from rats and humans-obtained from blood cultures at Kisangani University Hospital-was assessed with multilocus variable-number tandem repeat (VNTR) analysis (MLVA), multilocus sequence typing (MLST) and core-genome MLST (cgMLST). 1650 live-capture traps yielded 566 (34.3%) rats (95.6% Rattus norvegicus, 4.4% Rattus rattus); 46 (8.1%) of them carried Salmonella, of which 13 had more than one serotype. The most common serotypes were II.42:r:- (n = 18 rats), Kapemba (n = 12), Weltevreden and Typhimurium (n = 10, each), and Dublin (n = 8). Salmonella Typhimurium belonged to MLST ST19 (n = 7 rats) and the invasive ST313 (n = 3, isolated from deep organs but not from rectal content). Sixteen human S. Typhimurium isolates (all ST313) were available for comparison: MLVA and cgMLST revealed two distinct rat-human clusters involving both six human isolates, respectively, i.e. in total 12/16 human ST313 isolates. All ST313 Typhimurium isolates from rats and humans clustered with the ST313 Lineage 2 isolates and most were multidrug resistant; the remaining isolates from rats including S. Typhimurium ST19 were pan-susceptible.
Conclusion: The present study provides evidence of urban rats as potential reservoirs of S. Typhimurium ST313 in an iNTS endemic area in sub-Saharan Africa.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Stanaway JD, Parisi A, Sarkar K, Blacker BF, Reiner RC, Hay SI, et al. The global burden of non-typhoidal salmonella invasive disease: a systematic analysis for the Global Burden of Disease Study 2017. The Lancet Infectious Diseases. 2019;19: 1312–1324. doi: 10.1016/S1473-3099(19)30418-9 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

