Biosynthetic proteins targeting the SARS-CoV-2 spike as anti-virals
- PMID: 36067253
- PMCID: PMC9481167
- DOI: 10.1371/journal.ppat.1010799
Biosynthetic proteins targeting the SARS-CoV-2 spike as anti-virals
Abstract
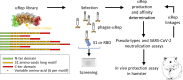
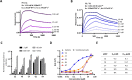
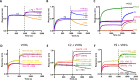
The binding of the SARS-CoV-2 spike to angiotensin-converting enzyme 2 (ACE2) promotes virus entry into the cell. Targeting this interaction represents a promising strategy to generate antivirals. By screening a phage-display library of biosynthetic protein sequences build on a rigid alpha-helicoidal HEAT-like scaffold (named αReps), we selected candidates recognizing the spike receptor binding domain (RBD). Two of them (F9 and C2) bind the RBD with affinities in the nM range, displaying neutralisation activity in vitro and recognizing distinct sites, F9 overlapping the ACE2 binding motif. The F9-C2 fusion protein and a trivalent αRep form (C2-foldon) display 0.1 nM affinities and EC50 of 8-18 nM for neutralization of SARS-CoV-2. In hamsters, F9-C2 instillation in the nasal cavity before or during infections effectively reduced the replication of a SARS-CoV-2 strain harbouring the D614G mutation in the nasal epithelium. Furthermore, F9-C2 and/or C2-foldon effectively neutralized SARS-CoV-2 variants (including delta and omicron variants) with EC50 values ranging from 13 to 32 nM. With their high stability and their high potency against SARS-CoV-2 variants, αReps provide a promising tool for SARS-CoV-2 therapeutics to target the nasal cavity and mitigate virus dissemination in the proximal environment.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Bryche B, St Albin A, Murri S, Lacôte S, Pulido C, Ar Gouilh M, et al. Massive transient damage of the olfactory epithelium associated with infection of sustentacular cells by SARS-CoV-2 in golden Syrian hamsters. Brain Behav Immun. 2020;89: 579–586. doi: 10.1016/j.bbi.2020.06.032 - DOI - PMC - PubMed
-
- Valdez-Cruz NA, García-Hernández E, Espitia C, Cobos-Marín L, Altamirano C, Bando-Campos CG, et al. Integrative overview of antibodies against SARS-CoV-2 and their possible applications in COVID-19 prophylaxis and treatment. Microb Cell Factories. 2021;20: 88. doi: 10.1186/s12934-021-01576-5 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

