Microbial functional diversity across biogeochemical provinces in the central Pacific Ocean
- PMID: 36067300
- PMCID: PMC9477243
- DOI: 10.1073/pnas.2200014119
Microbial functional diversity across biogeochemical provinces in the central Pacific Ocean
Abstract
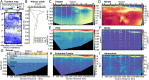
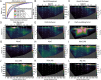
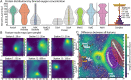
Enzymes catalyze key reactions within Earth's life-sustaining biogeochemical cycles. Here, we use metaproteomics to examine the enzymatic capabilities of the microbial community (0.2 to 3 µm) along a 5,000-km-long, 1-km-deep transect in the central Pacific Ocean. Eighty-five percent of total protein abundance was of bacterial origin, with Archaea contributing 1.6%. Over 2,000 functional KEGG Ontology (KO) groups were identified, yet only 25 KO groups contributed over half of the protein abundance, simultaneously indicating abundant key functions and a long tail of diverse functions. Vertical attenuation of individual proteins displayed stratification of nutrient transport, carbon utilization, and environmental stress. The microbial community also varied along horizontal scales, shaped by environmental features specific to the oligotrophic North Pacific Subtropical Gyre, the oxygen-depleted Eastern Tropical North Pacific, and nutrient-rich equatorial upwelling. Some of the most abundant proteins were associated with nitrification and C1 metabolisms, with observed interactions between these pathways. The oxidoreductases nitrite oxidoreductase (NxrAB), nitrite reductase (NirK), ammonia monooxygenase (AmoABC), manganese oxidase (MnxG), formate dehydrogenase (FdoGH and FDH), and carbon monoxide dehydrogenase (CoxLM) displayed distributions indicative of biogeochemical status such as oxidative or nutritional stress, with the potential to be more sensitive than chemical sensors. Enzymes that mediate transformations of atmospheric gases like CO, CO2, NO, methanethiol, and methylamines were most abundant in the upwelling region. We identified hot spots of biochemical transformation in the central Pacific Ocean, highlighted previously understudied metabolic pathways in the environment, and provided rich empirical data for biogeochemical models critical for forecasting ecosystem response to climate change.
Keywords: marine microbial ecology; mesopelagic; metaproteomics; methylotrophy; nitrification.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Falkowski P. G., Fenchel T., Delong E. F., The microbial engines that drive Earth’s biogeochemical cycles. Science 320, 1034–1039 (2008). - PubMed
-
- Morris R. M., et al. , Comparative metaproteomics reveals ocean-scale shifts in microbial nutrient utilization and energy transduction. ISME J. 4, 673–685 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

