Genomic surveillance of SARS-CoV-2 Omicron variants on a university campus
- PMID: 36068236
- PMCID: PMC9446629
- DOI: 10.1038/s41467-022-32786-z
Genomic surveillance of SARS-CoV-2 Omicron variants on a university campus
Abstract
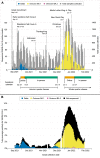
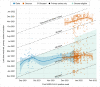
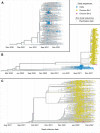
Novel variants continue to emerge in the SARS-CoV-2 pandemic. University testing programs may provide timely epidemiologic and genomic surveillance data to inform public health responses. We conducted testing from September 2021 to February 2022 in a university population under vaccination and indoor mask mandates. A total of 3,048 of 24,393 individuals tested positive for SARS-CoV-2 by RT-PCR; whole genome sequencing identified 209 Delta and 1,730 Omicron genomes of the 1,939 total sequenced. Compared to Delta, Omicron had a shorter median serial interval between genetically identical, symptomatic infections within households (2 versus 6 days, P = 0.021). Omicron also demonstrated a greater peak reproductive number (2.4 versus 1.8), and a 1.07 (95% confidence interval: 0.58, 1.57; P < 0.0001) higher mean cycle threshold value. Despite near universal vaccination and stringent mitigation measures, Omicron rapidly displaced the Delta variant to become the predominant viral strain and led to a surge in cases in a university population.
© 2022. The Author(s).
Conflict of interest statement
H.Y.C. reports consulting with Ellume, Pfizer, The Bill and Melinda Gates Foundation, Glaxo Smith Kline, and Merck. H.Y.C. received research funding from Gates Ventures, Sanofi Pasteur, and support and reagents from Ellume and Cepheid outside of the submitted work. G.S.G. received research grants and research support from the US National Institutes of Health, the University of Washington, the Bill & Melinda Gates Foundation, Gilead Sciences, Alere Technologies, Merck & Co., Janssen Pharmaceutica, Cerus Corporation, ViiV Healthcare, Bristol-Myers Squibb, Roche Molecular Systems, Abbott Molecular Diagnostics, and THERA Technologies/TaiMed Biologics, Inc, all outside of the submitted work. J.A.E. reports research support from Gates Ventures, AstraZeneca, GlaxoSmithKline, Merck, and Pfizer, and consulting with Sanofi Pasteur, AstraZeneca, Teva Pharmaceuticals, and Meissa Vaccines, outside of the submitted work. M.B. reports research support from Vir Biotechnology, GSK, Regeneron, Gilead Sciences, Janssen Pharmaceutica, Ridgeback, Merck, Gates Ventures, and consulting with Vir Biotechnology, Moderna, Helocyte, and Merck outside of the submitted work. The remaining authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

