Multimodal single cell sequencing implicates chromatin accessibility and genetic background in diabetic kidney disease progression
- PMID: 36068241
- PMCID: PMC9448792
- DOI: 10.1038/s41467-022-32972-z
Multimodal single cell sequencing implicates chromatin accessibility and genetic background in diabetic kidney disease progression
Abstract
The proximal tubule is a key regulator of kidney function and glucose metabolism. Diabetic kidney disease leads to proximal tubule injury and changes in chromatin accessibility that modify the activity of transcription factors involved in glucose metabolism and inflammation. Here we use single nucleus RNA and ATAC sequencing to show that diabetic kidney disease leads to reduced accessibility of glucocorticoid receptor binding sites and an injury-associated expression signature in the proximal tubule. We hypothesize that chromatin accessibility is regulated by genetic background and closely-intertwined with metabolic memory, which pre-programs the proximal tubule to respond differently to external stimuli. Glucocorticoid excess has long been known to increase risk for type 2 diabetes, which raises the possibility that glucocorticoid receptor inhibition may mitigate the adverse metabolic effects of diabetic kidney disease.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


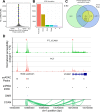

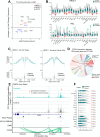


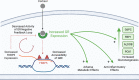
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

