Piezo1 regulates cholesterol biosynthesis to influence neural stem cell fate during brain development
- PMID: 36069933
- PMCID: PMC9458470
- DOI: 10.1085/jgp.202213084
Piezo1 regulates cholesterol biosynthesis to influence neural stem cell fate during brain development
Abstract
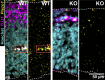
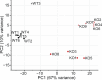
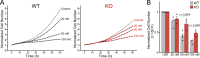
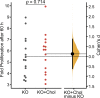
Mechanical forces and tissue mechanics influence the morphology of the developing brain, but the underlying molecular mechanisms have been elusive. Here, we examine the role of mechanotransduction in brain development by focusing on Piezo1, a mechanically activated ion channel. We find that Piezo1 deletion results in a thinner neuroepithelial layer, disrupts pseudostratification, and reduces neurogenesis in E10.5 mouse embryos. Proliferation and differentiation of Piezo1 knockout (KO) mouse neural stem cells (NSCs) isolated from E10.5 embryos are reduced in vitro compared to littermate WT NSCs. Transcriptome analysis of E10.5 Piezo1 KO brains reveals downregulation of the cholesterol biosynthesis superpathway, in which 16 genes, including Hmgcr, the gene encoding the rate-limiting enzyme of the cholesterol biosynthesis pathway, are downregulated by 1.5-fold or more. Consistent with this finding, membrane lipid composition is altered, and the cholesterol levels are reduced in Piezo1 KO NSCs. Cholesterol supplementation of Piezo1 KO NSCs partially rescues the phenotype in vitro. These findings demonstrate a role for Piezo1 in the neurodevelopmental process that modulates the quantity, quality, and organization of cells by influencing cellular cholesterol metabolism. Our study establishes a direct link in NSCs between PIEZO1, intracellular cholesterol levels, and neural development.
© 2022 Nourse et al.
Figures








References
-
- Albarrán-Juárez, J., Iring A., Wang S., Joseph S., Grimm M., Strilic B., Wettschureck N., Althoff T.F., and Offermanns S.. 2018. Piezo1 and Gq/G11 promote endothelial inflammation depending on flow pattern and integrin activation. J. Exp. Med. 215:2655–2672. 10.1084/jem.20180483 - DOI - PMC - PubMed
-
- Atcha, H., Jairaman A., Holt J.R., Meli V.S., Nagalla R.R., Veerasubramanian P.K., Brumm K.T., Lim H.E., Othy S., Cahalan M.D., et al. 2021a. Mechanically activated ion channel Piezo1 modulates macrophage polarization and stiffness sensing. Nat. Commun. 12:3256. 10.1038/s41467-021-23482-5 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

