Novel antigen-presenting cell imparts Treg-dependent tolerance to gut microbiota
- PMID: 36070798
- PMCID: PMC9605865
- DOI: 10.1038/s41586-022-05309-5
Novel antigen-presenting cell imparts Treg-dependent tolerance to gut microbiota
Abstract
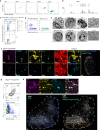
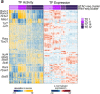
Establishing and maintaining tolerance to self-antigens or innocuous foreign antigens is vital for the preservation of organismal health. Within the thymus, medullary thymic epithelial cells (mTECs) expressing autoimmune regulator (AIRE) have a critical role in self-tolerance through deletion of autoreactive T cells and promotion of thymic regulatory T (Treg) cell development1-4. Within weeks of birth, a separate wave of Treg cell differentiation occurs in the periphery upon exposure to antigens derived from the diet and commensal microbiota5-8, yet the cell types responsible for the generation of peripheral Treg (pTreg) cells have not been identified. Here we describe the identification of a class of RORγt+ antigen-presenting cells called Thetis cells, with transcriptional features of both mTECs and dendritic cells, comprising four major sub-groups (TC I-TC IV). We uncover a developmental wave of Thetis cells within intestinal lymph nodes during a critical window in early life, coinciding with the wave of pTreg cell differentiation. Whereas TC I and TC III expressed the signature mTEC nuclear factor AIRE, TC IV lacked AIRE expression and was enriched for molecules required for pTreg generation, including the TGF-β-activating integrin αvβ8. Loss of either major histocompatibility complex class II (MHCII) or ITGB8 by Thetis cells led to a profound impairment in intestinal pTreg differentiation, with ensuing colitis. By contrast, MHCII expression by RORγt+ group 3 innate lymphoid cells (ILC3) and classical dendritic cells was neither sufficient nor required for pTreg generation, further implicating TC IV as the tolerogenic RORγt+ antigen-presenting cell with an essential function in early life. Our studies reveal parallel pathways for the establishment of tolerance to self and foreign antigens in the thymus and periphery, respectively, marked by the involvement of shared cellular and transcriptional programmes.
© 2022. The Author(s).
Conflict of interest statement
M.v.d.B. has received research support and stock options from Seres Therapeutics and stock options from Notch Therapeutics and Pluto Therapeutics; he has received royalties from Wolters Kluwer; has consulted, received honorarium from or participated in advisory boards for Seres Therapeutics, WindMIL Therapeutics, Rheos Medicines, Merck & Co., Magenta Therapeutics, Frazier Healthcare Partners, Nektar Therapeutics, Notch Therapeutics, Forty Seven, Priothera, Ceramedix, Lygenesis, Pluto Therapeutics, GlaskoSmithKline, Da Volterra, Vor BioPharma, Novartis (spouse), Synthekine (spouse) and Beigene (spouse); he has intellectual property licensed to Seres Therapeutics and Juno Therapeutics; and holds a fiduciary role on the Foundation Board of DKMS (a nonprofit organization). A.Y.R. is a member of the scientific advisory board and has equity in Surface Oncology and RAPT Therapeutics, and holds intellectual property licensed to Takeda, which is not related to the content of this study.
Figures













Comment in
-
How regulatory T cells are primed to aid tolerance of gut bacteria.Nature. 2022 Oct;610(7933):638-640. doi: 10.1038/d41586-022-03368-2. Nature. 2022. PMID: 36280729 No abstract available.
-
Novel "Thetis" antigen-presenting cells promote early life gut immune tolerance.Immunol Cell Biol. 2023 Jan;101(1):16-19. doi: 10.1111/imcb.12605. Epub 2022 Nov 22. Immunol Cell Biol. 2023. PMID: 36333957 No abstract available.
-
An embarrassment of riches: RORγt+ antigen-presenting cells in peripheral tolerance.Immunity. 2022 Nov 8;55(11):1978-1980. doi: 10.1016/j.immuni.2022.10.009. Immunity. 2022. PMID: 36351372 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

