ILC3s select microbiota-specific regulatory T cells to establish tolerance in the gut
- PMID: 36071169
- PMCID: PMC9613541
- DOI: 10.1038/s41586-022-05141-x
ILC3s select microbiota-specific regulatory T cells to establish tolerance in the gut
Abstract
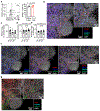
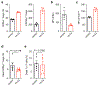
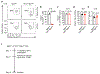
Microbial colonization of the mammalian intestine elicits inflammatory or tolerogenic T cell responses, but the mechanisms controlling these distinct outcomes remain poorly understood, and accumulating evidence indicates that aberrant immunity to intestinal microbiota is causally associated with infectious, inflammatory and malignant diseases1-8. Here we define a critical pathway controlling the fate of inflammatory versus tolerogenic T cells that respond to the microbiota and express the transcription factor RORγt. We profiled all RORγt+ immune cells at single-cell resolution from the intestine-draining lymph nodes of mice and reveal a dominant presence of T regulatory (Treg) cells and lymphoid tissue inducer-like group 3 innate lymphoid cells (ILC3s), which co-localize at interfollicular regions. These ILC3s are distinct from extrathymic AIRE-expressing cells, abundantly express major histocompatibility complex class II, and are necessary and sufficient to promote microbiota-specific RORγt+ Treg cells and prevent their expansion as inflammatory T helper 17 cells. This occurs through ILC3-mediated antigen presentation, αV integrin and competition for interleukin-2. Finally, single-cell analyses suggest that interactions between ILC3s and RORγt+ Treg cells are impaired in inflammatory bowel disease. Our results define a paradigm whereby ILC3s select for antigen-specific RORγt+ Treg cells, and against T helper 17 cells, to establish immune tolerance to the microbiota and intestinal health.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare no other competing interests.
Figures












Comment in
-
How regulatory T cells are primed to aid tolerance of gut bacteria.Nature. 2022 Oct;610(7933):638-640. doi: 10.1038/d41586-022-03368-2. Nature. 2022. PMID: 36280729 No abstract available.
-
An embarrassment of riches: RORγt+ antigen-presenting cells in peripheral tolerance.Immunity. 2022 Nov 8;55(11):1978-1980. doi: 10.1016/j.immuni.2022.10.009. Immunity. 2022. PMID: 36351372 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- 110199/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- R01 CA274534/CA/NCI NIH HHS/United States
- R01 AI162936/AI/NIAID NIH HHS/United States
- R21 CA249274/CA/NCI NIH HHS/United States
- R01 AI145989/AI/NIAID NIH HHS/United States
- R01 AI123368/AI/NIAID NIH HHS/United States
- R01 DK126871/DK/NIDDK NIH HHS/United States
- R35 CA210088/CA/NCI NIH HHS/United States
- P30 ES002109/ES/NIEHS NIH HHS/United States
- UL1 TR002384/TR/NCATS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 AI143842/AI/NIAID NIH HHS/United States
- U01 AI095608/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

